The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons
- PMID: 26950095
- PMCID: PMC4817229
- DOI: 10.1038/ng.3526
The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons
Abstract
To connect human biology to fish biomedical models, we sequenced the genome of spotted gar (Lepisosteus oculatus), whose lineage diverged from teleosts before teleost genome duplication (TGD). The slowly evolving gar genome has conserved in content and size many entire chromosomes from bony vertebrate ancestors. Gar bridges teleosts to tetrapods by illuminating the evolution of immunity, mineralization and development (mediated, for example, by Hox, ParaHox and microRNA genes). Numerous conserved noncoding elements (CNEs; often cis regulatory) undetectable in direct human-teleost comparisons become apparent using gar: functional studies uncovered conserved roles for such cryptic CNEs, facilitating annotation of sequences identified in human genome-wide association studies. Transcriptomic analyses showed that the sums of expression domains and expression levels for duplicated teleost genes often approximate the patterns and levels of expression for gar genes, consistent with subfunctionalization. The gar genome provides a resource for understanding evolution after genome duplication, the origin of vertebrate genomes and the function of human regulatory sequences.
Figures

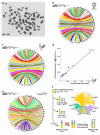
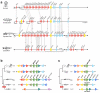
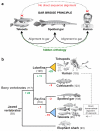

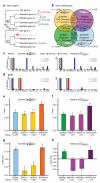
Comment in
-
The gar is a fish... is a bird... is a mammal?Nat Genet. 2016 Apr;48(4):344-5. doi: 10.1038/ng.3532. Nat Genet. 2016. PMID: 27023772
-
Corrigendum: The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons.Nat Genet. 2016 May 27;48(6):700. doi: 10.1038/ng0616-700c. Nat Genet. 2016. PMID: 27230688 No abstract available.
-
Subfunctionalization versus neofunctionalization after whole-genome duplication.Nat Genet. 2018 Jul;50(7):908-909. doi: 10.1038/s41588-018-0162-4. Nat Genet. 2018. PMID: 29955176 No abstract available.
References
-
- Nelson J S. Fishes of the world. 4th edn. John Wiley; 2006.
-
- Patton EE, Mathers ME, Schartl M. Generating and analyzing fish models of melanoma. Methods in cell biology. 2011;105:339–366. - PubMed
-
- Lee O, Green JM, Tyler CR. Transgenic fish systems and their application in ecotoxicology. Crit Rev Toxicol. 2015;45:124–141. - PubMed
Methods-only references
-
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. - PubMed
Publication types
MeSH terms
Grants and funding
- R24 OD010922/OD/NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R24 OD011199/OD/NIH HHS/United States
- R01 GM079492/GM/NIGMS NIH HHS/United States
- R24 OD01119004/OD/NIH HHS/United States
- R01 AI057559/AI/NIAID NIH HHS/United States
- R01 RR020833/RR/NCRR NIH HHS/United States
- R21 GM079492/GM/NIGMS NIH HHS/United States
- WT095908/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- WT098051/WT_/Wellcome Trust/United Kingdom
- U54 HG03067/HG/NHGRI NIH HHS/United States
- 268513/ERC_/European Research Council/International
- T32 HD055164/HD/NICHD NIH HHS/United States
- R01 OD011116/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

