A New Nanobody-Based Biosensor to Study Endogenous PARP1 In Vitro and in Live Human Cells
- PMID: 26950694
- PMCID: PMC4780744
- DOI: 10.1371/journal.pone.0151041
A New Nanobody-Based Biosensor to Study Endogenous PARP1 In Vitro and in Live Human Cells
Abstract
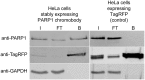
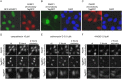
Poly(ADP-ribose) polymerase 1 (PARP1) is a key player in DNA repair, genomic stability and cell survival and it emerges as a highly relevant target for cancer therapies. To deepen our understanding of PARP biology and mechanisms of action of PARP1-targeting anti-cancer compounds, we generated a novel PARP1-affinity reagent, active both in vitro and in live cells. This PARP1-biosensor is based on a PARP1-specific single-domain antibody fragment (~ 15 kDa), termed nanobody, which recognizes the N-terminus of human PARP1 with nanomolar affinity. In proteomic approaches, immobilized PARP1 nanobody facilitates quantitative immunoprecipitation of functional, endogenous PARP1 from cellular lysates. For cellular studies, we engineered an intracellularly functional PARP1 chromobody by combining the nanobody coding sequence with a fluorescent protein sequence. By following the chromobody signal, we were for the first time able to monitor the recruitment of endogenous PARP1 to DNA damage sites in live cells. Moreover, tracing of the sub-nuclear translocation of the chromobody signal upon treatment of human cells with chemical substances enables real-time profiling of active compounds in high content imaging. Due to its ability to perform as a biosensor at the endogenous level of the PARP1 enzyme, the novel PARP1 nanobody is a unique and versatile tool for basic and applied studies of PARP1 biology and DNA repair.
Conflict of interest statement
Figures






References
-
- Kameshita I, Matsuda Z, Taniguchi T, Shizuta Y. Poly (ADP-Ribose) synthetase. Separation and identification of three proteolytic fragments as the substrate-binding domain, the DNA-binding domain, and the automodification domain. The Journal of biological chemistry. 1984;259(8):4770–6. Epub 1984/04/25. . - PubMed
-
- de Murcia G, Menissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends in biochemical sciences. 1994;19(4):172–6. Epub 1994/04/01. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

