Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease
- PMID: 26950749
- PMCID: PMC4824656
- DOI: 10.1038/nbt.3513
Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease
Abstract
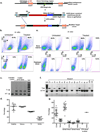
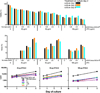
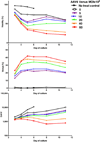
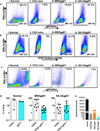
Gene therapy with genetically modified human CD34(+) hematopoietic stem and progenitor cells (HSPCs) may be safer using targeted integration (TI) of transgenes into a genomic 'safe harbor' site rather than random viral integration. We demonstrate that temporally optimized delivery of zinc finger nuclease mRNA via electroporation and adeno-associated virus (AAV) 6 delivery of donor constructs in human HSPCs approaches clinically relevant levels of TI into the AAVS1 safe harbor locus. Up to 58% Venus(+) HSPCs with 6-16% human cell marking were observed following engraftment into mice. In HSPCs from patients with X-linked chronic granulomatous disease (X-CGD), caused by mutations in the gp91phox subunit of the NADPH oxidase, TI of a gp91phox transgene into AAVS1 resulted in ∼15% gp91phox expression and increased NADPH oxidase activity in ex vivo-derived neutrophils. In mice transplanted with corrected HSPCs, 4-11% of human cells in the bone marrow expressed gp91phox. This method for TI into AAVS1 may be broadly applicable to correction of other monogenic diseases.
Figures
References
-
- Biffi A, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. - PubMed
-
- Cavazzana-Calvo M, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. - PubMed
-
- Hacein-Bey-Abina S, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. The New England journal of medicine. 2003;348:255–256. - PubMed
-
- Stein S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nature medicine. 2010;16:198–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

