The gut microbiome of healthy Japanese and its microbial and functional uniqueness
- PMID: 26951067
- PMCID: PMC4833420
- DOI: 10.1093/dnares/dsw002
The gut microbiome of healthy Japanese and its microbial and functional uniqueness
Abstract
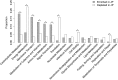
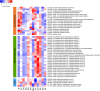
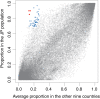
The human gut microbiome has profound influences on the host's health largely through its interference with various intestinal functions. As recent studies have suggested diversity in the human gut microbiome among human populations, it will be interesting to analyse how gut microbiome is correlated with geographical, cultural, and traditional differences. The Japanese people are known to have several characteristic features such as eating a variety of traditional foods and exhibiting a low BMI and long life span. In this study, we analysed gut microbiomes of the Japanese by comparing the metagenomic data obtained from 106 Japanese individuals with those from 11 other nations. We found that the composition of the Japanese gut microbiome showed more abundant in the phylum Actinobacteria, in particular in the genus Bifidobacterium, than other nations. Regarding the microbial functions, those of carbohydrate metabolism were overrepresented with a concurrent decrease in those for replication and repair, and cell motility. The remarkable low prevalence of genes for methanogenesis with a significant depletion of the archaeon Methanobrevibacter smithii and enrichment of acetogenesis genes in the Japanese gut microbiome compared with others suggested a difference in the hydrogen metabolism pathway in the gut between them. It thus seems that the gut microbiome of the Japanese is considerably different from those of other populations, which cannot be simply explained by diet alone. We postulate possible existence of hitherto unknown factors contributing to the population-level diversity in human gut microbiomes.
Keywords: Japanese; gut; metagenome; microbiome.
© The Author 2016. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures


References
-
- Qin J., Li Y., Cai Z. et al. . 2012, A metagenome-wide association study of gut microbiota in type 2 diabetes, Nature, 490, 55–60. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

