SUMOylation Regulates Growth Factor Independence 1 in Transcriptional Control and Hematopoiesis
- PMID: 26951200
- PMCID: PMC4859688
- DOI: 10.1128/MCB.01001-15
SUMOylation Regulates Growth Factor Independence 1 in Transcriptional Control and Hematopoiesis
Abstract
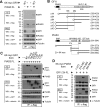
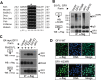
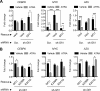
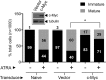
Cell fate specification requires precise coordination of transcription factors and their regulators to achieve fidelity and flexibility in lineage allocation. The transcriptional repressor growth factor independence 1 (GFI1) is comprised of conserved Snail/Slug/Gfi1 (SNAG) and zinc finger motifs separated by a linker region poorly conserved with GFI1B, its closest homolog. Moreover, GFI1 and GFI1B coordinate distinct developmental fates in hematopoiesis, suggesting that their functional differences may derive from structures within their linkers. We show a binding interface between the GFI1 linker and the SP-RING domain of PIAS3, an E3-SUMO (small ubiquitin-related modifier) ligase. The PIAS3 binding region in GFI1 contains a conserved type I SUMOylation consensus element, centered on lysine-239 (K239). In silico prediction algorithms identify K239 as the only high-probability site for SUMO modification. We show that GFI1 is modified by SUMO at K239. SUMOylation-resistant derivatives of GFI1 fail to complement Gfi1 depletion phenotypes in zebrafish primitive erythropoiesis and granulocytic differentiation in cultured human cells. LSD1/CoREST recruitment and MYC repression by GFI1 are profoundly impaired for SUMOylation-resistant GFI1 derivatives, while enforced expression of MYC blocks granulocytic differentiation. These findings suggest that SUMOylation within the GFI1 linker favors LSD1/CoREST recruitment and MYC repression to govern hematopoietic differentiation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Growth Factor Independence 1B-Mediated Transcriptional Repression and Lineage Allocation Require Lysine-Specific Demethylase 1-Dependent Recruitment of the BHC Complex.Mol Cell Biol. 2019 Jun 13;39(13):e00020-19. doi: 10.1128/MCB.00020-19. Print 2019 Jul 1. Mol Cell Biol. 2019. PMID: 30988160 Free PMC article.
-
PIASγ controls stability and facilitates SUMO-2 conjugation to CoREST family of transcriptional co-repressors.Biochem J. 2018 Apr 23;475(8):1441-1454. doi: 10.1042/BCJ20170983. Biochem J. 2018. PMID: 29555846
-
GFI1 functions in transcriptional control and cell fate determination require SNAG domain methylation to recruit LSD1.Biochem J. 2016 Oct 1;473(19):3355-69. doi: 10.1042/BCJ20160558. Epub 2016 Aug 1. Biochem J. 2016. PMID: 27480105
-
Gfi1 and Gfi1b: key regulators of hematopoiesis.Leukemia. 2010 Nov;24(11):1834-43. doi: 10.1038/leu.2010.195. Epub 2010 Sep 23. Leukemia. 2010. PMID: 20861919 Review.
-
From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation.Blood. 2015 Dec 10;126(24):2561-9. doi: 10.1182/blood-2015-06-655043. Epub 2015 Oct 7. Blood. 2015. PMID: 26447191 Free PMC article. Review.
Cited by
-
SUMO and Transcriptional Regulation: The Lessons of Large-Scale Proteomic, Modifomic and Genomic Studies.Molecules. 2021 Feb 5;26(4):828. doi: 10.3390/molecules26040828. Molecules. 2021. PMID: 33562565 Free PMC article. Review.
-
SENP1 promotes MCL pathogenesis through regulating JAK-STAT5 pathway and SOCS2 expression.Cell Death Discov. 2021 Jul 26;7(1):192. doi: 10.1038/s41420-021-00578-x. Cell Death Discov. 2021. PMID: 34312374 Free PMC article.
-
Multifaceted Actions of GFI1 and GFI1B in Hematopoietic Stem Cell Self-Renewal and Lineage Commitment.Front Genet. 2020 Oct 26;11:591099. doi: 10.3389/fgene.2020.591099. eCollection 2020. Front Genet. 2020. PMID: 33193732 Free PMC article. Review.
-
Biallelic Loss of Function Variants in SENP7 Cause Immunodeficiency with Neurologic and Muscular Phenotypes.J Pediatr. 2024 Nov;274:114180. doi: 10.1016/j.jpeds.2024.114180. Epub 2024 Jul 5. J Pediatr. 2024. PMID: 38972567
-
The scaffolding function of LSD1/KDM1A reinforces a negative feedback loop to repress stem cell gene expression during primitive hematopoiesis.iScience. 2022 Dec 5;26(1):105737. doi: 10.1016/j.isci.2022.105737. eCollection 2023 Jan 20. iScience. 2022. PMID: 36594016 Free PMC article.
References
-
- Gilks CB, Bear SE, Grimes HL, Tsichlis PN. 1993. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol 13:1759–1768. doi:10.1128/MCB.13.3.1759. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
