Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases
- PMID: 26952246
- PMCID: PMC4786772
- DOI: 10.1038/ncomms10849
Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases
Abstract
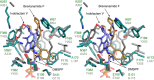
Prenylation reactions play crucial roles in controlling the activities of biomolecules. Bacterial prenyltransferases, TleC from Streptomyces blastmyceticus and MpnD from Marinactinospora thermotolerans, catalyse the 'reverse' prenylation of (-)-indolactam V at the C-7 position of the indole ring with geranyl pyrophosphate or dimethylallyl pyrophosphate, to produce lyngbyatoxin or pendolmycin, respectively. Using in vitro analyses, here we show that both TleC and MpnD exhibit relaxed substrate specificities and accept various chain lengths (C5-C25) of the prenyl donors. Comparisons of the crystal structures and their ternary complexes with (-)-indolactam V and dimethylallyl S-thiophosphate revealed the intimate structural details of the enzyme-catalysed 'reverse' prenylation reactions and identified the active-site residues governing the selection of the substrates. Furthermore, structure-based enzyme engineering successfully altered the preference for the prenyl chain length of the substrates, as well as the regio- and stereo-selectivities of the prenylation reactions, to produce a series of unnatural novel indolactams.
Figures




Similar articles
-
Characterization of a single gene cluster responsible for methylpendolmycin and pendolmycin biosynthesis in the deep sea bacterium Marinactinospora thermotolerans.Chembiochem. 2012 Mar 5;13(4):547-52. doi: 10.1002/cbic.201100700. Epub 2012 Feb 23. Chembiochem. 2012. PMID: 22362652
-
Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593.J Bacteriol. 2010 Jun;192(11):2839-51. doi: 10.1128/JB.01557-09. Epub 2010 Mar 26. J Bacteriol. 2010. PMID: 20348259 Free PMC article.
-
Structural basis for non-genuine phenolic acceptor substrate specificity of Streptomyces roseochromogenes prenyltransferase CloQ from the ABBA/PT-barrel superfamily.PLoS One. 2017 Mar 29;12(3):e0174665. doi: 10.1371/journal.pone.0174665. eCollection 2017. PLoS One. 2017. PMID: 28355308 Free PMC article.
-
Biosynthetic studies on teleocidins in Streptomyces.J Antibiot (Tokyo). 2018 Sep;71(9):763-768. doi: 10.1038/s41429-018-0069-4. Epub 2018 Jun 14. J Antibiot (Tokyo). 2018. PMID: 29904063 Review.
-
Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives.Curr Med Chem. 2009;16(2):218-31. doi: 10.2174/092986709787002772. Curr Med Chem. 2009. PMID: 19149573 Review.
Cited by
-
Biochemical characterization of a cyanobactin arginine-N-prenylase from the autumnalamide biosynthetic pathway.Chem Commun (Camb). 2022 Oct 27;58(86):12054-12057. doi: 10.1039/d2cc01799g. Chem Commun (Camb). 2022. PMID: 36193595 Free PMC article.
-
Microbial soluble aromatic prenyltransferases for engineered biosynthesis.Synth Syst Biotechnol. 2021 Mar 15;6(2):51-62. doi: 10.1016/j.synbio.2021.02.001. eCollection 2021 Jun. Synth Syst Biotechnol. 2021. PMID: 33778178 Free PMC article.
-
Synthetic production of prenylated naringenins in yeast using promiscuous microbial prenyltransferases.Metab Eng Commun. 2021 Mar 19;12:e00169. doi: 10.1016/j.mec.2021.e00169. eCollection 2021 Jun. Metab Eng Commun. 2021. PMID: 33868922 Free PMC article.
-
C7-Prenylation of Tryptophan-Containing Cyclic Dipeptides by 7-Dimethylallyl Tryptophan Synthase Significantly Increases the Anticancer and Antimicrobial Activities.Molecules. 2020 Aug 12;25(16):3676. doi: 10.3390/molecules25163676. Molecules. 2020. PMID: 32806659 Free PMC article.
-
Prenylation of dimeric cyclo-L-Trp-L-Trp by the promiscuous cyclo-L-Trp-L-Ala prenyltransferase EchPT1.Appl Microbiol Biotechnol. 2023 Nov;107(22):6887-6895. doi: 10.1007/s00253-023-12773-0. Epub 2023 Sep 15. Appl Microbiol Biotechnol. 2023. PMID: 37713115 Free PMC article.
References
-
- Nakagawa K. et al. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature 468, 117–121 (2010). - PubMed
-
- Sakagami Y., Yoshida M., Isogai A. & Suzuki A. Peptidal sex hormones inducing conjugation tube formation in compatible mating-type cells of Tremella mesenterica. Science 212, 1525–1527 (1981). - PubMed
-
- Okada M. et al. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1, 23–24 (2005). - PubMed
-
- Brown M. S. & Goldstein J. L. Protein prenylation. Mad bet for Rab. Nature 366, 14–15 (1993). - PubMed
-
- Cui C. B. et al. Tryprostatins A and B, novel mammalian cell cycle inhibitors produced by Aspergillus fumigatus. J. Antibiot. 48, 1382–1384 (1995). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

