The Paired-box protein PAX-3 regulates the choice between lateral and ventral epidermal cell fates in C. elegans
- PMID: 26953187
- PMCID: PMC4846358
- DOI: 10.1016/j.ydbio.2016.03.002
The Paired-box protein PAX-3 regulates the choice between lateral and ventral epidermal cell fates in C. elegans
Abstract
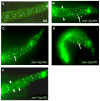
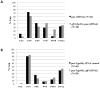
The development of the single cell layer skin or hypodermis of Caenorhabditis elegans is an excellent model for understanding cell fate specification and differentiation. Early in C. elegans embryogenesis, six rows of hypodermal cells adopt dorsal, lateral or ventral fates that go on to display distinct behaviors during larval life. Several transcription factors are known that function in specifying these major hypodermal cell fates, but our knowledge of the specification of these cell types is sparse, particularly in the case of the ventral hypodermal cells, which become Vulval Precursor Cells and form the vulval opening in response to extracellular signals. Previously, the gene pvl-4 was identified in a screen for mutants with defects in vulval development. We found by whole genome sequencing that pvl-4 is the Paired-box gene pax-3, which encodes the sole PAX-3 transcription factor homolog in C. elegans. pax-3 mutants show embryonic and larval lethality, and body morphology abnormalities indicative of hypodermal cell defects. We report that pax-3 is expressed in ventral P cells and their descendants during embryogenesis and early larval stages, and that in pax-3 reduction-of-function animals the ventral P cells undergo a cell fate transformation and express several markers of the lateral seam cell fate. Furthermore, forced expression of pax-3 in the lateral hypodermal cells causes them to lose expression of seam cell markers. We propose that pax-3 functions in the ventral hypodermal cells to prevent these cells from adopting the lateral seam cell fate. pax-3 represents the first gene required for specification solely of the ventral hypodermal fate in C. elegans providing insights into cell type diversification.
Keywords: C. elegans; Differentiation; Epidermis; Fate specification; Gene expression; PAX.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no actual or potential conflict of interest including financial, personal or other relationships with other people or organizations that could inappropriately influence, or be perceived to influence, the work reported here.
Figures






References
-
- Andachi Y. Caenorhabditis elegans T-box genes tbx-9 and tbx-8 are required for formation of hypodermis and body-wall muscle in embryogenesis. Genes Cells. 2004;9:331–344. - PubMed
-
- Aspöck G, Kagoshima H, Niklaus G, Bürglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–923. - PubMed
-
- Blake JA, Thomas M, Thompson JA, White R, Ziman M. Perplexing Pax: from puzzle to paradigm. Dev Dyn. 2008;237:2791–2803. - PubMed
-
- Blake JA, Ziman MR. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141:737–751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

