CotG-Like Modular Proteins Are Common among Spore-Forming Bacilli
- PMID: 26953338
- PMCID: PMC4859607
- DOI: 10.1128/JB.00023-16
CotG-Like Modular Proteins Are Common among Spore-Forming Bacilli
Abstract
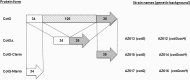
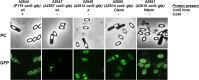
CotG is an abundant protein initially identified as an outer component of the Bacillus subtilis spore coat. It has an unusual structure characterized by several repeats of positively charged amino acids that are probably the outcome of multiple rounds of gene elongation events in an ancestral minigene. CotG is not highly conserved, and its orthologues are present in only two Bacillus and two Geobacillus species. In B. subtilis, CotG is the target of extensive phosphorylation by a still unidentified enzyme and has a role in the assembly of some outer coat proteins. We report now that most spore-forming bacilli contain a protein not homologous to CotG of B. subtilis but sharing a central "modular" region defined by a pronounced positive charge and randomly coiled tandem repeats. Conservation of the structural features in most spore-forming bacilli suggests a relevant role for the CotG-like protein family in the structure and function of the bacterial endospore. To expand our knowledge on the role of CotG, we dissected the B. subtilis protein by constructing deletion mutants that express specific regions of the protein and observed that they have different roles in the assembly of other coat proteins and in spore germination.
Importance: CotG of B. subtilis is not highly conserved in the Bacillus genus; however, a CotG-like protein with a modular structure and chemical features similar to those of CotG is common in spore-forming bacilli, at least when CotH is also present. The conservation of CotG-like features when CotH is present suggests that the two proteins act together and may have a relevant role in the structure and function of the bacterial endospore. Dissection of the modular composition of CotG of B. subtilis by constructing mutants that express only some of the modules has allowed a first characterization of CotG modules and will be the basis for a more detailed functional analysis.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
A protein phosphorylation module patterns the Bacillus subtilis spore outer coat.Mol Microbiol. 2020 Dec;114(6):934-951. doi: 10.1111/mmi.14562. Epub 2020 Sep 12. Mol Microbiol. 2020. PMID: 32592201 Free PMC article.
-
CotG Mediates Spore Surface Permeability in Bacillus subtilis.mBio. 2022 Dec 20;13(6):e0276022. doi: 10.1128/mbio.02760-22. Epub 2022 Nov 10. mBio. 2022. PMID: 36354330 Free PMC article.
-
Organization and evolution of the cotG and cotH genes of Bacillus subtilis.J Bacteriol. 2011 Dec;193(23):6664-73. doi: 10.1128/JB.06121-11. Epub 2011 Oct 7. J Bacteriol. 2011. PMID: 21984783 Free PMC article.
-
[Identification and characterization of the outermost layer of Bacillus subtilis spores].Yakugaku Zasshi. 2012;132(8):919-24. doi: 10.1248/yakushi.132.919. Yakugaku Zasshi. 2012. PMID: 22864350 Review. Japanese.
-
Assembly and genetics of spore protective structures.Cell Mol Life Sci. 2002 Mar;59(3):434-44. doi: 10.1007/s00018-002-8436-4. Cell Mol Life Sci. 2002. PMID: 11964122 Free PMC article. Review.
Cited by
-
A protein phosphorylation module patterns the Bacillus subtilis spore outer coat.Mol Microbiol. 2020 Dec;114(6):934-951. doi: 10.1111/mmi.14562. Epub 2020 Sep 12. Mol Microbiol. 2020. PMID: 32592201 Free PMC article.
-
CotG Mediates Spore Surface Permeability in Bacillus subtilis.mBio. 2022 Dec 20;13(6):e0276022. doi: 10.1128/mbio.02760-22. Epub 2022 Nov 10. mBio. 2022. PMID: 36354330 Free PMC article.
-
Role of the Spore Coat Proteins CotA and CotB, and the Spore Surface Protein CDIF630_02480, on the Surface Distribution of Exosporium Proteins in Clostridioides difficile 630 Spores.Microorganisms. 2022 Sep 27;10(10):1918. doi: 10.3390/microorganisms10101918. Microorganisms. 2022. PMID: 36296193 Free PMC article.
-
Spectroscopic Characterization and Biological Effects of 1-Oxo-bisabolone-rich Pulicaria burchardii Hutch. subsp. burchardii Essential Oil Against Viruses, Bacteria, and Spore Germination.Plants (Basel). 2024 Dec 29;14(1):68. doi: 10.3390/plants14010068. Plants (Basel). 2024. PMID: 39795328 Free PMC article.
-
Fructose-Induced Impairment of Liver and Skeletal Muscle Metabolism Is Prevented by Administration of Shouchella clausii Spores by Preserving Mitochondrial Function and Insulin Sensitivity.Mol Nutr Food Res. 2025 Jun;69(11):e70063. doi: 10.1002/mnfr.70063. Epub 2025 Apr 10. Mol Nutr Food Res. 2025. PMID: 40207597 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

