Population Structure and Antimicrobial Resistance Profiles of Streptococcus suis Serotype 2 Sequence Type 25 Strains
- PMID: 26954687
- PMCID: PMC4783015
- DOI: 10.1371/journal.pone.0150908
Population Structure and Antimicrobial Resistance Profiles of Streptococcus suis Serotype 2 Sequence Type 25 Strains
Abstract
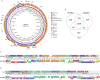
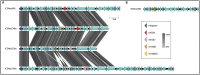
Strains of serotype 2 Streptococcus suis are responsible for swine and human infections. Different serotype 2 genetic backgrounds have been defined using multilocus sequence typing (MLST). However, little is known about the genetic diversity within each MLST sequence type (ST). Here, we used whole-genome sequencing to test the hypothesis that S. suis serotype 2 strains of the ST25 lineage are genetically heterogeneous. We evaluated 51 serotype 2 ST25 S. suis strains isolated from diseased pigs and humans in Canada, the United States of America, and Thailand. Whole-genome sequencing revealed numerous large-scale rearrangements in the ST25 genome, compared to the genomes of ST1 and ST28 S. suis strains, which result, among other changes, in disruption of a pilus island locus. We report that recombination and lateral gene transfer contribute to ST25 genetic diversity. Phylogenetic analysis identified two main and distinct Thai and North American clades grouping most strains investigated. These clades also possessed distinct patterns of antimicrobial resistance genes, which correlated with acquisition of different integrative and conjugative elements (ICEs). Some of these ICEs were found to be integrated at a recombination hot spot, previously identified as the site of integration of the 89K pathogenicity island in serotype 2 ST7 S. suis strains. Our results highlight the limitations of MLST for phylogenetic analysis of S. suis, and the importance of lateral gene transfer and recombination as drivers of diversity in this swine pathogen and zoonotic agent.
Conflict of interest statement
Figures

References
-
- Gottschalk M. Streptococcosis In: Zimmerman JJ KL, Ramirez A, Schwartz JK, Stevenson G, editor. Diseases of swine. Mes, Iowa: Wiley Publishers; 2011. p. 841–55.
-
- Suankratay C, Intalapaporn P, Nunthapisud P, Arunyingmongkol K, Wilde H. Streptococcus suis meningitis in Thailand. Southeast Asian J Trop Med Pub Health. 2004;35(4):868–76. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

