Improving gastric cancer preclinical studies using diverse in vitro and in vivo model systems
- PMID: 26955870
- PMCID: PMC4784390
- DOI: 10.1186/s12885-016-2232-2
Improving gastric cancer preclinical studies using diverse in vitro and in vivo model systems
Abstract
Background: "Biomarker-driven targeted therapy," the practice of tailoring patients' treatment to the expression/activity levels of disease-specific genes/proteins, remains challenging. For example, while the anti-ERBB2 monoclonal antibody, trastuzumab, was first developed using well-characterized, diverse in vitro breast cancer models (and is now a standard adjuvant therapy for ERBB2-positive breast cancer patients), trastuzumab approval for ERBB2-positive gastric cancer was largely based on preclinical studies of a single cell line, NCI-N87. Ensuing clinical trials revealed only modest patient efficacy, and many ERBB2-positive gastric cancer (GC) patients failed to respond at all (i.e., were inherently recalcitrant), or succumbed to acquired resistance.
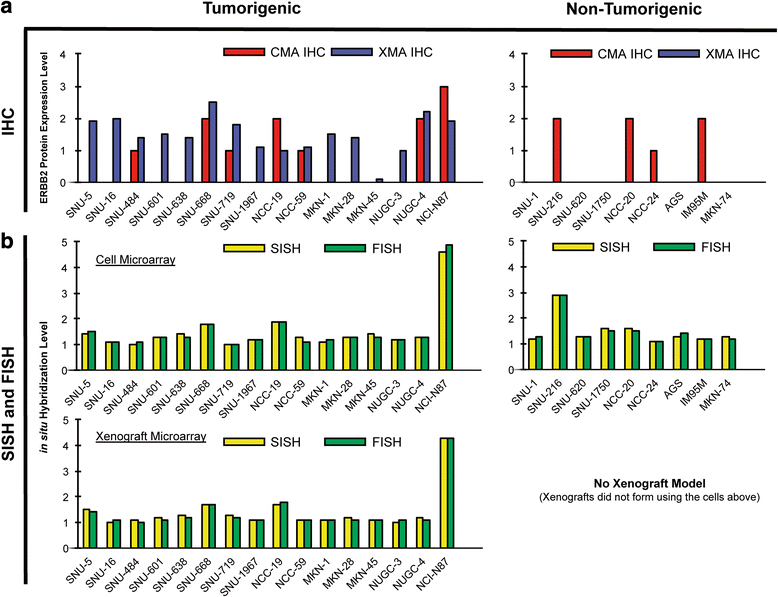
Method: To assess mechanisms underlying GC insensitivity to ERBB2 therapies, we established a diverse panel of GC cells, differing in ERBB2 expression levels, for comprehensive in vitro and in vivo characterization. For higher throughput assays of ERBB2 DNA and protein levels, we compared the concordance of various laboratory quantification methods, including those of in vitro and in vivo genetic anomalies (FISH and SISH) and xenograft protein expression (Western blot vs. IHC), of both cell and xenograft (tissue-sectioned) microarrays.
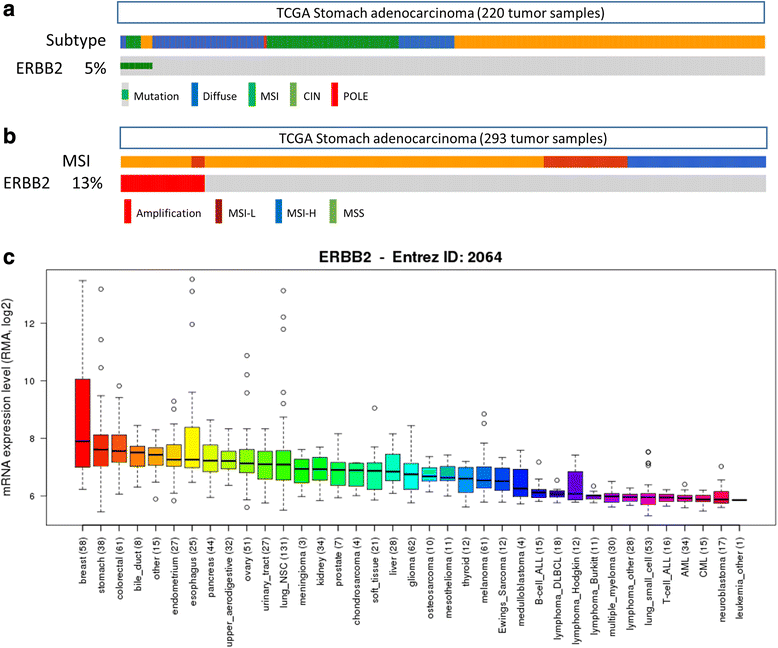
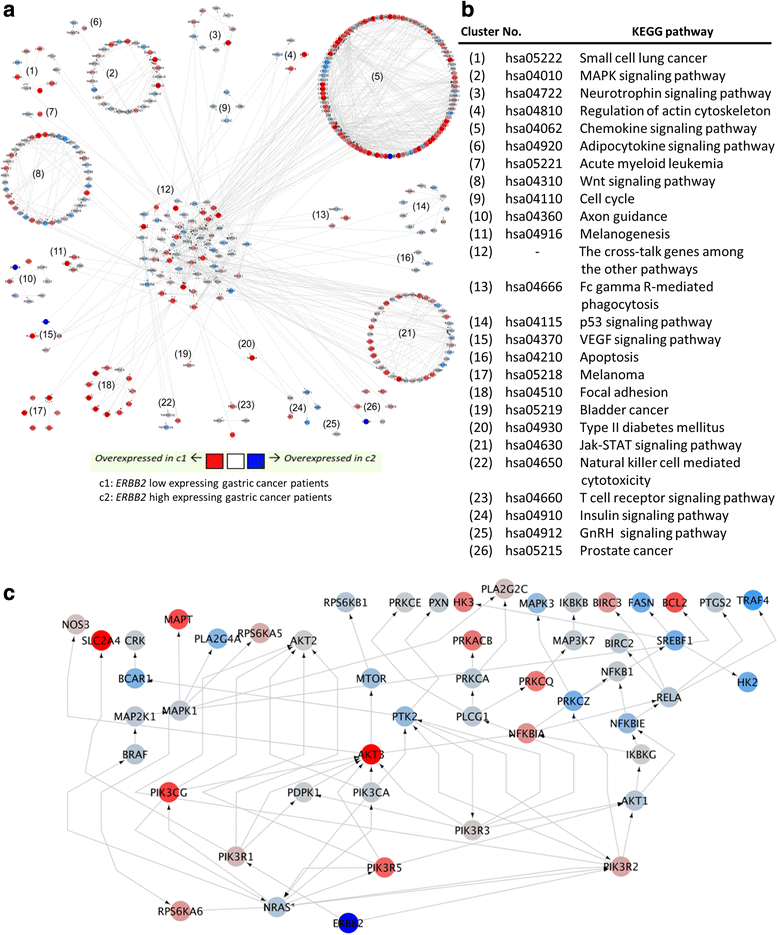
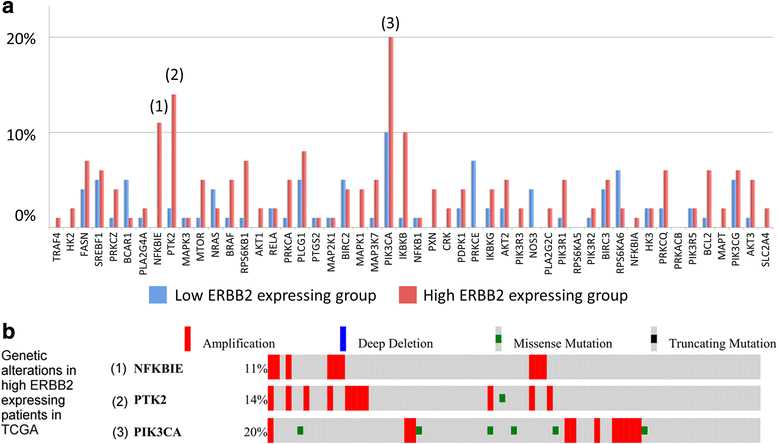
Results: The biomarker assessment methods strongly agreed, as did correlation between RNA and protein expression. However, although ERBB2 genomic anomalies showed good in vitro vs. in vivo correlation, we observed striking differences in protein expression between cultured cells and mouse xenografts (even within the same GC cell type). Via our unique pathway analysis, we delineated a signaling network, in addition to specific pathways/biological processes, emanating from the ERBB2 signaling cascade, as a potential useful target of clinical treatment. Integrated analysis of public data from gastric tumors revealed frequent (10 - 20 %) amplification of the genes NFKBIE, PTK2, and PIK3CA, each of which resides in an ERBB2-derived subpathway network.
Conclusion: Our comprehensive bioinformatics analyses of highly heterogeneous cancer cells, combined with tumor "omics" profiles, can optimally characterize the expression patterns and activity of specific tumor biomarkers. Subsequent in vitro and in vivo validation, of specific disease biomarkers (using multiple methodologies), can improve prediction of patient stratification according to drug response or nonresponse.
Figures

Similar articles
-
Establishment of patient-derived gastric cancer xenografts: a useful tool for preclinical evaluation of targeted therapies involving alterations in HER-2, MET and FGFR2 signaling pathways.BMC Cancer. 2017 Mar 14;17(1):191. doi: 10.1186/s12885-017-3177-9. BMC Cancer. 2017. PMID: 28292264 Free PMC article.
-
Dual PI3K/mTOR inhibitor BEZ235 exerts extensive antitumor activity in HER2-positive gastric cancer.BMC Cancer. 2015 Nov 11;15:894. doi: 10.1186/s12885-015-1900-y. BMC Cancer. 2015. PMID: 26560145 Free PMC article.
-
Evaluation of Trastuzumab Anti-Tumor Efficacy and its Correlation with HER-2 Status in Patient-Derived Gastric Adenocarcinoma Xenograft Models.Pathol Oncol Res. 2015 Sep;21(4):947-55. doi: 10.1007/s12253-015-9909-8. Epub 2015 Mar 9. Pathol Oncol Res. 2015. PMID: 25749810 Free PMC article.
-
Cardiotoxicity of ErbB2-targeted therapies and its impact on drug development, a spotlight on trastuzumab.Expert Opin Drug Metab Toxicol. 2017 Jul;13(7):755-766. doi: 10.1080/17425255.2017.1337746. Epub 2017 Jun 9. Expert Opin Drug Metab Toxicol. 2017. PMID: 28571477 Review.
-
HER-2 positive gastric cancer: Current targeted treatments.Int J Biol Macromol. 2024 Aug;274(Pt 1):133247. doi: 10.1016/j.ijbiomac.2024.133247. Epub 2024 Jun 19. Int J Biol Macromol. 2024. PMID: 38906351 Review.
Cited by
-
RHOA in Gastric Cancer: Functional Roles and Therapeutic Potential.Front Genet. 2019 May 15;10:438. doi: 10.3389/fgene.2019.00438. eCollection 2019. Front Genet. 2019. PMID: 31156701 Free PMC article. Review.
-
RHOA protein expression correlates with clinical features in gastric cancer: a systematic review and meta-analysis.BMC Cancer. 2022 Jul 19;22(1):798. doi: 10.1186/s12885-022-09904-7. BMC Cancer. 2022. PMID: 35854253 Free PMC article.
-
A Deep Learning Model for Cell Growth Inhibition IC50 Prediction and Its Application for Gastric Cancer Patients.Int J Mol Sci. 2019 Dec 12;20(24):6276. doi: 10.3390/ijms20246276. Int J Mol Sci. 2019. PMID: 31842404 Free PMC article.
-
Databases and tools for constructing signal transduction networks in cancer.BMB Rep. 2017 Jan;50(1):12-19. doi: 10.5483/bmbrep.2017.50.1.135. BMB Rep. 2017. PMID: 27502015 Free PMC article. Review.
-
Gene Regulation and Targeted Therapy in Gastric Cancer Peritoneal Metastasis: Radiological Findings from Dual Energy CT and PET/CT.J Vis Exp. 2018 Jan 22;(131):56526. doi: 10.3791/56526. J Vis Exp. 2018. PMID: 29443079 Free PMC article.
References
-
- Singh RK, Singh DP, Tiwari SP, Mohapatra TM. Targeted therapies for cancer treatment. J Pharm Res. 2011;4(2):312–16.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

