The genomic architecture of NLRP7 is Alu rich and predisposes to disease-associated large deletions
- PMID: 26956250
- PMCID: PMC5027696
- DOI: 10.1038/ejhg.2016.9
The genomic architecture of NLRP7 is Alu rich and predisposes to disease-associated large deletions
Erratum in
-
The genomic architecture of NLRP7 is Alu rich and predisposes to disease-associated large deletions.Eur J Hum Genet. 2016 Oct;24(10):1516. doi: 10.1038/ejhg.2016.96. Eur J Hum Genet. 2016. PMID: 27628567 Free PMC article. No abstract available.
Abstract
NLRP7 is a major gene responsible for recurrent hydatidiform moles. Here, we report 11 novel NLRP7 protein truncating variants, of which five deletions of more than 1-kb. We analyzed the transcriptional consequences of four variants. We demonstrate that one large homozygous deletion removes NLRP7 transcription start site and results in the complete absence of its transcripts in a patient in good health besides her reproductive problem. This observation strengthens existing data on the requirement of NLRP7 only for female reproduction. We show that two other variants affecting the splice acceptor of exon 6 lead to its in-frame skipping while another variant affecting the splice donor site of exon 9 leads to an in-frame insertion of 54 amino acids. Our characterization of the deletion breakpoints demonstrated that most of the breakpoints occurred within Alu repeats and the deletions were most likely mediated by microhomology events. Our data define a hotspot of Alu instability and deletions in intron 5 with six different breakpoints and rearrangements. Analysis of NLRP7 genomic sequences for repetitive elements demonstrated that Alu repeats represent 48% of its intronic sequences and these repeats seem to have been inserted into the common NLRP2/7 primate ancestor before its duplication into two genes.
Figures


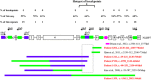

References
-
- Savage P, Williams J, Wong SL et al: The demographics of molar pregnancies in England and Wales from 2000–2009. J Reprod Med 2010; 55: 341–345. - PubMed
-
- Grimes DA: Epidemiology of gestational trophoblastic disease. Am J Obstet Gynecol 1984; 150: 309–318. - PubMed
-
- Eagles N, Sebire NJ, Short D, Savage PM, Seckl MJ, Fisher RA: Risk of recurrent molar pregnancies following complete and partial hydatidiform moles. Hum Reprod 2015; 30: 2055–2063. - PubMed
-
- Sebire NJ, Fisher RA, Foskett M, Rees H, Seckl MJ, Newlands ES: Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. BJOG 2003; 110: 22–26. - PubMed
-
- Horn LC, Kowalzik J, Bilek K, Richter CE, Einenkel J: Clinicopathologic characteristics and subsequent pregnancy outcome in 139 complete hydatidiform moles. Eur J Obstet Gynecol Reprod Biol 2006; 128: 10–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

