The Diversification of Evolutionarily Conserved MAPK Cascades Correlates with the Evolution of Fungal Species and Development of Lifestyles
- PMID: 26957028
- PMCID: PMC5381651
- DOI: 10.1093/gbe/evw051
The Diversification of Evolutionarily Conserved MAPK Cascades Correlates with the Evolution of Fungal Species and Development of Lifestyles
Abstract
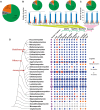
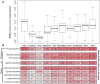
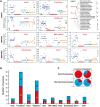
The fungal kingdom displays an extraordinary diversity of lifestyles, developmental processes, and ecological niches. The MAPK (mitogen-activated protein kinase) cascade consists of interlinked MAPKKK, MAPKK, and MAPK, and collectively such cascades play pivotal roles in cellular regulation in fungi. However, the mechanism by which evolutionarily conserved MAPK cascades regulate diverse output responses in fungi remains unknown. Here we identified the full complement of MAPK cascade components from 231 fungal species encompassing 9 fungal phyla. Using the largest data set to date, we found that MAPK family members could have two ancestors, while MAPKK and MAPKKK family members could have only one ancestor. The current MAPK, MAPKK, and MAPKKK subfamilies resulted from duplications and subsequent subfunctionalization during the emergence of the fungal kingdom. However, the gene structure diversification and gene expansion and loss have resulted in significant diversity in fungal MAPK cascades, correlating with the evolution of fungal species and lifestyles. In particular, a distinct evolutionary trajectory of MAPK cascades was identified in single-celled fungi in the Saccharomycetes. All MAPK, MAPKK, and MAPKKK subfamilies expanded in the Saccharomycetes; genes encoding MAPK cascade components have a similar exon-intron structure in this class that differs from those in other fungi.
Keywords: MAPK cascades; evolutionary diversity; fungi; gene duplication; gene family evolution; gene loss.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. 2004. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415. - PubMed
-
- Basse CW, Steinberg G. 2004. Ustilago maydis, model system for analysis of the molecular basis of fungal pathogenicity. Mol Plant Pathol. 5:83–92. - PubMed
-
- Clarke DL, Woodlee GL, McClelland CM, Seymour TS, Wickes BL. 2001. The Cryptococcus neoformans STE11alpha gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol Microbiol. 40:200–213. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

