Drosophila O-GlcNAcase Deletion Globally Perturbs Chromatin O-GlcNAcylation
- PMID: 26957542
- PMCID: PMC4858994
- DOI: 10.1074/jbc.M115.704783
Drosophila O-GlcNAcase Deletion Globally Perturbs Chromatin O-GlcNAcylation
Abstract
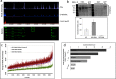
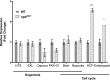
Gene expression during Drosophila development is subject to regulation by the Polycomb (Pc), Trithorax (Trx), and Compass chromatin modifier complexes. O-GlcNAc transferase (OGT/SXC) is essential for Pc repression suggesting that the O-GlcNAcylation of proteins plays a key role in regulating development. OGT transfers O-GlcNAc onto serine and threonine residues in intrinsically disordered domains of key transcriptional regulators; O-GlcNAcase (OGA) removes the modification. To pinpoint genomic regions that are regulated by O-GlcNAc levels, we performed ChIP-chip and microarray analysis after OGT or OGA RNAi knockdown in S2 cells. After OGA RNAi, we observed a genome-wide increase in the intensity of most O-GlcNAc-occupied regions including genes linked to cell cycle, ubiquitin, and steroid response. In contrast, O-GlcNAc levels were strikingly insensitive to OGA RNAi at sites of polycomb repression such as the Hox and NK homeobox gene clusters. Microarray analysis suggested that altered O-GlcNAc cycling perturbed the expression of genes associated with morphogenesis and cell cycle regulation. We then produced a viable null allele of oga (oga(del.1)) in Drosophila allowing visualization of altered O-GlcNAc cycling on polytene chromosomes. We found that trithorax (TRX), absent small or homeotic discs 1 (ASH1), and Compass member SET1 histone methyltransferases were O-GlcNAc-modified in oga(del.1) mutants. The oga(del.1) mutants displayed altered expression of a distinct set of cell cycle-related genes. Our results show that the loss of OGA in Drosophila globally impacts the epigenetic machinery allowing O-GlcNAc accumulation on RNA polymerase II and numerous chromatin factors including TRX, ASH1, and SET1.
Keywords: Ash1; Drosophila; O-GlcNAcylation; O-linked N-acetylglucosamine (O-GlcNAc); RNA polymerase II; SET1; Trithorax; cell cycle; chromatin; polycomb.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Iovino N., Ciabrelli F., and Cavalli G. (2013) PRC2 controls Drosophila oocyte cell fate by repressing cell cycle genes. Dev. Cell 26, 431–439 - PubMed
-
- Martinez A. M., and Cavalli G. (2006) The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5, 1189–1197 - PubMed
-
- Schuettengruber B., Martinez A. M., Iovino N., and Cavalli G. (2011) Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

