Human symbionts inject and neutralize antibacterial toxins to persist in the gut
- PMID: 26957597
- PMCID: PMC4822603
- DOI: 10.1073/pnas.1525637113
Human symbionts inject and neutralize antibacterial toxins to persist in the gut
Abstract
The human gut microbiome is a dynamic and densely populated microbial community that can provide important benefits to its host. Cooperation and competition for nutrients among its constituents only partially explain community composition and interpersonal variation. Notably, certain human-associated Bacteroidetes--one of two major phyla in the gut--also encode machinery for contact-dependent interbacterial antagonism, but its impact within gut microbial communities remains unknown. Here we report that prominent human gut symbionts persist in the gut through continuous attack on their immediate neighbors. Our analysis of just one of the hundreds of species in these communities reveals 12 candidate antibacterial effector loci that can exist in 32 combinations. Through the use of secretome studies, in vitro bacterial interaction assays and multiple mouse models, we uncover strain-specific effector/immunity repertoires that can predict interbacterial interactions in vitro and in vivo, and find that some of these strains avoid contact-dependent killing by accumulating immunity genes to effectors that they do not encode. Effector transmission rates in live animals can exceed 1 billion events per minute per gram of colonic contents, and multiphylum communities of human gut commensals can partially protect sensitive strains from these attacks. Together, these results suggest that gut microbes can determine their interactions through direct contact. An understanding of the strategies human gut symbionts have evolved to target other members of this community may provide new approaches for microbiome manipulation.
Keywords: gut microbiome; microbial ecology; symbiosis; type VI secretion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



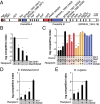
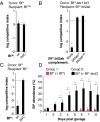
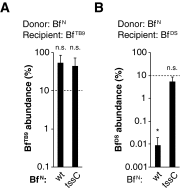
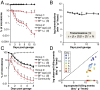
References
-
- Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM101209/GM/NIGMS NIH HHS/United States
- DP2 GM105456/GM/NIGMS NIH HHS/United States
- Canadian Institutes of Health Research/Canada
- OD008440/OD/NIH HHS/United States
- GM103574/GM/NIGMS NIH HHS/United States
- R35 GM118038/GM/NIGMS NIH HHS/United States
- DP2 OD008440/OD/NIH HHS/United States
- Howard Hughes Medical Institute/United States
- GM108657/GM/NIGMS NIH HHS/United States
- AI080609/AI/NIAID NIH HHS/United States
- GM105456/GM/NIGMS NIH HHS/United States
- GM101209/GM/NIGMS NIH HHS/United States
- R01 GM103574/GM/NIGMS NIH HHS/United States
- R01 AI080609/AI/NIAID NIH HHS/United States
- R01 GM108657/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

