Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq
- PMID: 26958077
- PMCID: PMC4782572
- DOI: 10.1186/s13068-016-0471-8
Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq
Abstract
Background: Arundo donax L. (Poaceae) is considered one of the most promising energy crops in the Mediterranean region because of its high biomass yield and low input requirements, but to date no information on its transcriptional responses to water stress is available.
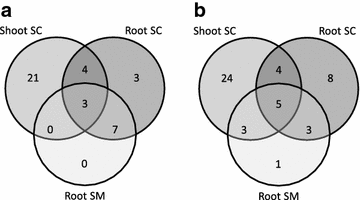
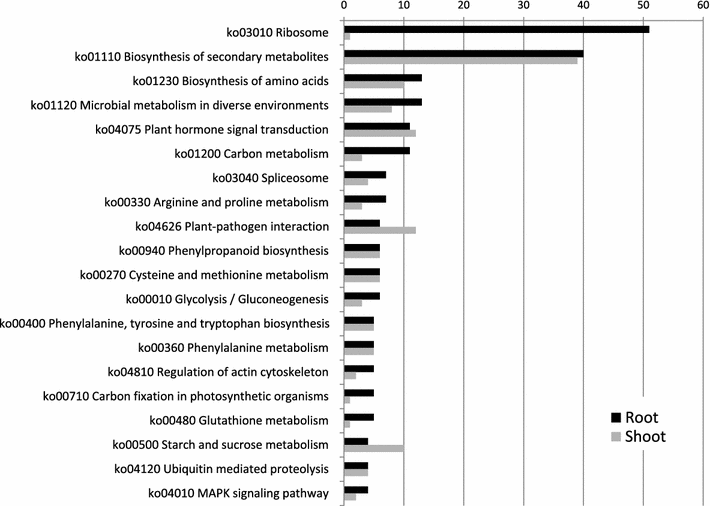
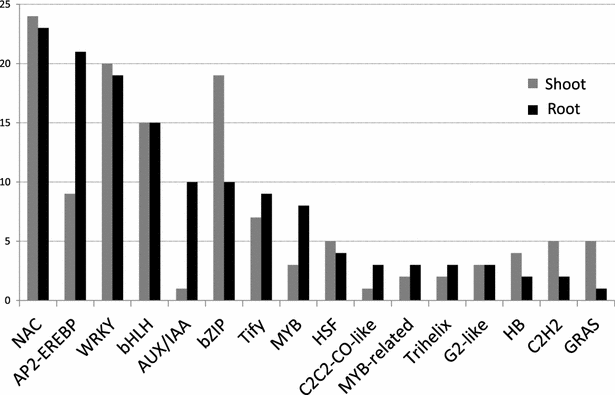
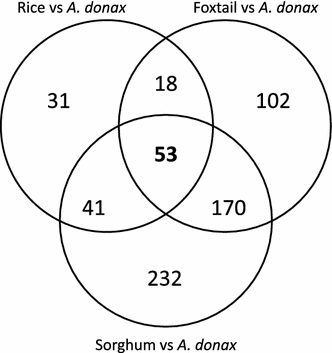
Results: We obtained by Illumina-based RNA-seq the whole root and shoot transcriptomes of young A. donax plants subjected to osmotic/water stress with 10 and 20 % polyethylene glycol (PEG; 3 biological replicates/organ/condition corresponding to 18 RNA-Seq libraries), and identified a total of 3034 differentially expressed genes. Blast-based mining of stress-related genes indicated the higher responsivity of roots compared to shoots at the early stages of water stress especially under the milder PEG treatment, with a majority of genes responsive to salt, oxidative, and dehydration stress. Analysis of gene ontology terms underlined the qualitatively different responses between root and shoot tissues. Among the most significantly enriched metabolic pathways identified using a Fisher's exact test with FDR correction, a crucial role was played in both shoots and roots by genes involved in the signaling cascade of abscisic acid. We further identified relatively large organ-specific differences in the patterns of drought-related transcription factor AP2-EREBP, AUX/IAA, MYB, bZIP, C2H2, and GRAS families, which may underlie the transcriptional reprogramming differences between organs. Through comparative analyses with major Poaceae species based on Blast, we finally identified a set of 53 orthologs that can be considered as a core of evolutionary conserved genes important to mediate water stress responses in the family.
Conclusions: This study provides the first characterization of A. donax transcriptome in response to water stress, thus shedding novel light at the molecular level on the mechanisms of stress response and adaptation in this emerging bioenergy species. The inventory of early-responsive genes to water stress identified could constitute useful markers of the physiological status of A. donax and be a basis for the improvement of its productivity under water limitation. The full water-stressed A. donax transcriptome is available for Blast-based homology searches through a dedicated web server (http://ecogenomics.fmach.it/arundo/).
Keywords: Abscisic acid (ABA) signaling; Arundo donax; Conserved drought-responsive genes; Osmoregulatory proline metabolism; Poaceae; RNA-Seq; Transcription factors; Water stress.
Figures


Similar articles
-
Transcriptional response of giant reed (Arundo donax L.) low ecotype to long-term salt stress by unigene-based RNAseq.Phytochemistry. 2020 Sep;177:112436. doi: 10.1016/j.phytochem.2020.112436. Epub 2020 Jun 18. Phytochemistry. 2020. PMID: 32563719
-
Global leaf and root transcriptome in response to cadmium reveals tolerance mechanisms in Arundo donax L.BMC Genomics. 2022 Jun 8;23(1):427. doi: 10.1186/s12864-022-08605-6. BMC Genomics. 2022. PMID: 35672691 Free PMC article.
-
Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery.BMC Plant Biol. 2019 Jan 11;19(1):23. doi: 10.1186/s12870-018-1626-5. BMC Plant Biol. 2019. PMID: 30634906 Free PMC article.
-
Arundo donax L.: a non-food crop for bioenergy and bio-compound production.Biotechnol Adv. 2014 Dec;32(8):1535-49. doi: 10.1016/j.biotechadv.2014.10.006. Epub 2014 Oct 16. Biotechnol Adv. 2014. PMID: 25457226 Review.
-
Arundo donax L., a candidate for phytomanaging water and soils contaminated by trace elements and producing plant-based feedstock. A review.Int J Phytoremediation. 2014;16(7-12):982-1017. doi: 10.1080/15226514.2013.810580. Int J Phytoremediation. 2014. PMID: 24933898 Review.
Cited by
-
Genetic Improvement of Arundo donax L.: Opportunities and Challenges.Plants (Basel). 2020 Nov 16;9(11):1584. doi: 10.3390/plants9111584. Plants (Basel). 2020. PMID: 33207586 Free PMC article. Review.
-
De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock.Biotechnol Biofuels. 2017 May 30;10:138. doi: 10.1186/s13068-017-0828-7. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28572841 Free PMC article.
-
In Planta Recapitulation of Isoprene Synthase Evolution from Ocimene Synthases.Mol Biol Evol. 2017 Oct 1;34(10):2583-2599. doi: 10.1093/molbev/msx178. Mol Biol Evol. 2017. PMID: 28637270 Free PMC article.
-
In silico identification and characterization of a diverse subset of conserved microRNAs in bioenergy crop Arundo donax L.Sci Rep. 2018 Nov 12;8(1):16667. doi: 10.1038/s41598-018-34982-8. Sci Rep. 2018. PMID: 30420632 Free PMC article.
-
Drought and salinity induced changes in ecophysiology and proteomic profile of Parthenium hysterophorus.PLoS One. 2017 Sep 27;12(9):e0185118. doi: 10.1371/journal.pone.0185118. eCollection 2017. PLoS One. 2017. PMID: 28953916 Free PMC article.
References
-
- Acres G. Alcoholic Fuels. Platin Met Rev. 2007;51:34–35. doi: 10.1595/147106707X170009. - DOI
-
- Lewandowski I, Scurlock JMO, Lindvall E, Christou M. The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenerg. 2003;25:335–361. doi: 10.1016/S0961-9534(03)00030-8. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

