Contrasting Association Results between Existing PheWAS Phenotype Definition Methods and Five Validated Electronic Phenotypes
- PMID: 26958218
- PMCID: PMC4765620
Contrasting Association Results between Existing PheWAS Phenotype Definition Methods and Five Validated Electronic Phenotypes
Abstract
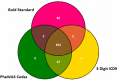
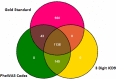
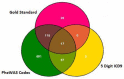
Phenome-Wide Association Studies (PheWAS) comprehensively investigate the association between genetic variation and a wide array of outcome traits. Electronic health record (EHR) based PheWAS uses various abstractions of International Classification of Diseases, Ninth Revision (ICD-9) codes to identify case/control status for diagnoses that are used as the phenotypic variables. However, there have not been comparisons within a PheWAS between results from high quality derived phenotypes and high-throughput but potentially inaccurate use of ICD-9 codes for case/control definition. For this study we first developed a group of high quality algorithms for five phenotypes. Next we evaluated the association of these "gold standard" phenotypes and 4,636,178 genetic variants with minor allele frequency > 0.01 and compared the results from high-throughput associations at the 3 digit, 5 digit, and PheWAS codes for defining case/control status. We found that certain diseases contained similar patient populations across phenotyping methods but had differences in PheWAS.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
