Dynamical Allocation of Cellular Resources as an Optimal Control Problem: Novel Insights into Microbial Growth Strategies
- PMID: 26958858
- PMCID: PMC4784908
- DOI: 10.1371/journal.pcbi.1004802
Dynamical Allocation of Cellular Resources as an Optimal Control Problem: Novel Insights into Microbial Growth Strategies
Abstract
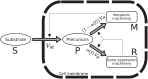
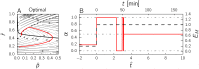
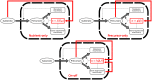
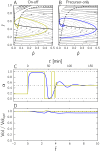
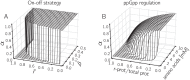
Microbial physiology exhibits growth laws that relate the macromolecular composition of the cell to the growth rate. Recent work has shown that these empirical regularities can be derived from coarse-grained models of resource allocation. While these studies focus on steady-state growth, such conditions are rarely found in natural habitats, where microorganisms are continually challenged by environmental fluctuations. The aim of this paper is to extend the study of microbial growth strategies to dynamical environments, using a self-replicator model. We formulate dynamical growth maximization as an optimal control problem that can be solved using Pontryagin's Maximum Principle. We compare this theoretical gold standard with different possible implementations of growth control in bacterial cells. We find that simple control strategies enabling growth-rate maximization at steady state are suboptimal for transitions from one growth regime to another, for example when shifting bacterial cells to a medium supporting a higher growth rate. A near-optimal control strategy in dynamical conditions is shown to require information on several, rather than a single physiological variable. Interestingly, this strategy has structural analogies with the regulation of ribosomal protein synthesis by ppGpp in the enterobacterium Escherichia coli. It involves sensing a mismatch between precursor and ribosome concentrations, as well as the adjustment of ribosome synthesis in a switch-like manner. Our results show how the capability of regulatory systems to integrate information about several physiological variables is critical for optimizing growth in a changing environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Schaechter M, Ingraham JL, Neidhardt FC. Microbe. ASM Press; 2006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

