Retroviral Integrase: Then and Now
- PMID: 26958915
- PMCID: PMC4810031
- DOI: 10.1146/annurev-virology-100114-055043
Retroviral Integrase: Then and Now
Abstract
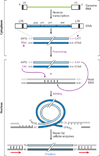
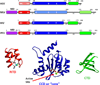
The retroviral integrases are virally encoded, specialized recombinases that catalyze the insertion of viral DNA into the host cell's DNA, a process that is essential for virus propagation. We have learned a great deal since the existence of an integrated form of retroviral DNA (the provirus) was first proposed by Howard Temin in 1964. Initial studies focused on the genetics and biochemistry of avian and murine virus DNA integration, but the pace of discovery increased substantially with advances in technology, and an influx of investigators focused on the human immunodeficiency virus. We begin with a brief account of the scientific landscape in which some of the earliest discoveries were made, and summarize research that led to our current understanding of the biochemistry of integration. A more detailed account of recent analyses of integrase structure follows, as they have provided valuable insights into enzyme function and raised important new questions.
Keywords: core dimer; intasome; integrase; prophage; provirus; reaching dimer.
Figures


References
-
- Wollman E, Wollman E. Les «phases» des bactériophages (facteurs lysogènes) C.R. Soc. Biol. 1937;124:931–934.
-
- Jacob F, Wollman E. Sexuality and the Genetics of Bacteria. New York: Academic; 1961.
-
- Campbell A. Episomes. Adv. Genet. 1961;11:101–145.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

