Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets
- PMID: 26960398
- PMCID: PMC4995776
- DOI: 10.1158/1078-0432.CCR-15-2946
Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets
Abstract
Purpose: Pulmonary large cell neuroendocrine carcinoma (LCNEC) is a highly aggressive neoplasm, whose biologic relationship to small cell lung carcinoma (SCLC) versus non-SCLC (NSCLC) remains unclear, contributing to uncertainty regarding optimal clinical management. To clarify these relationships, we analyzed genomic alterations in LCNEC compared with other major lung carcinoma types.
Experimental design: LCNEC (n = 45) tumor/normal pairs underwent targeted next-generation sequencing of 241 cancer genes by Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) platform and comprehensive histologic, immunohistochemical, and clinical analysis. Genomic data were compared with MSK-IMPACT analysis of other lung carcinoma histologies (n = 242).
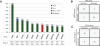
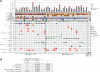
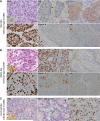
Results: Commonly altered genes in LCNEC included TP53 (78%), RB1 (38%), STK11 (33%), KEAP1 (31%), and KRAS (22%). Genomic profiles segregated LCNEC into 2 major and 1 minor subsets: SCLC-like (n = 18), characterized by TP53+RB1 co-mutation/loss and other SCLC-type alterations, including MYCL amplification; NSCLC-like (n = 25), characterized by the lack of coaltered TP53+RB1 and nearly universal occurrence of NSCLC-type mutations (STK11, KRAS, and KEAP1); and carcinoid-like (n = 2), characterized by MEN1 mutations and low mutation burden. SCLC-like and NSCLC-like subsets revealed several clinicopathologic differences, including higher proliferative activity in SCLC-like tumors (P < 0.0001) and exclusive adenocarcinoma-type differentiation marker expression in NSCLC-like tumors (P = 0.005). While exhibiting predominant similarity with lung adenocarcinoma, NSCLC-like LCNEC harbored several distinctive genomic alterations, including more frequent mutations in NOTCH family genes (28%), implicated as key regulators of neuroendocrine differentiation.
Conclusions: LCNEC is a biologically heterogeneous group of tumors, comprising distinct subsets with genomic signatures of SCLC, NSCLC (predominantly adenocarcinoma), and rarely, highly proliferative carcinoids. Recognition of these subsets may inform the classification and management of LCNEC patients. Clin Cancer Res; 22(14); 3618-29. ©2016 AACR.
©2016 American Association for Cancer Research.
Figures
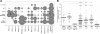

References
-
- Travis WD, Linnoila RI, Tsokos MG, Hitchcock CL, Cutler GB, Jr, Nieman L, et al. Neuroendocrine tumors of the lung with proposed criteria for large-cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical, and flow cytometric study of 35 cases. Am J Surg Pathol. 1991;15:529–53. - PubMed
-
- Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG, editors. WHO Classification of tumours of the lung, pleura, thymus and heart. 4th ed. IARC Press; Lyon, France: 2015. - PubMed
-
- McDowell EM, Wilson TS, Trump BF. Atypical endocrine tumors of the lung. Arch Pathol Lab Med. 1981;105:20–8. - PubMed
-
- Rekhtman N. Neuroendocrine tumors of the lung: an update. Arch Pathol Lab Med. 2010;134:1628–38. - PubMed
-
- Pelosi G, Hiroshima K, Mino-Kenudson M. Controversial issues and new discoveries in lung neuroendocrine tumors. Diagn Histopathol. 2014;20:392–7.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

