Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD
- PMID: 26961875
- PMCID: PMC4861456
- DOI: 10.1074/jbc.C115.699918
Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD
Abstract
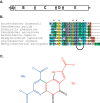
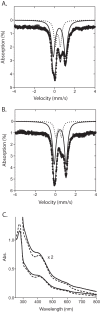
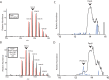
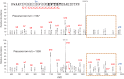
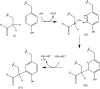
The radical S-adenosylmethionine (SAM) protein PqqE is predicted to function in the pyrroloquinoline quinone (PQQ) biosynthetic pathway via catalysis of carbon-carbon bond formation between a glutamate and tyrosine side chain within the small peptide substrate PqqA. We report here that PqqE activity is dependent on the accessory protein PqqD, which was recently shown to bind PqqA tightly. In addition, PqqE activity in vitro requires the presence of a flavodoxin- and flavodoxin reductase-based reduction system, with other reductants leading to an uncoupled cleavage of the co-substrate SAM. These results indicate that PqqE, in conjunction with PqqD, carries out the first step in PQQ biosynthesis: a radical-mediated formation of a new carbon-carbon bond between two amino acid side chains on PqqA.
Keywords: PQQ; S-adenosylmethionine (SAM); SPASM domain; flavoprotein; iron-sulfur protein; peptides; post-translational modification (PTM); radical SAM.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
References
-
- Duine J. A. (1999) The PQQ story. J. Biosci. Bioeng. 88, 231–236 - PubMed
-
- van Kleef M. A., and Duine J. A. (1988) A search for intermediates in the bacterial biosynthesis of PQQ. Biofactors 1, 297–302 - PubMed
-
- Magnusson O. T., Toyama H., Saeki M., Rojas A., Reed J. C., Liddington R. C., Klinman J. P., and Schwarzenbacher R. (2004) Quinone biogenesis: structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl. Acad. Sci. U.S.A. 101, 7913–7918 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

