Molecular Basis and Consequences of the Cytochrome c-tRNA Interaction
- PMID: 26961879
- PMCID: PMC4858987
- DOI: 10.1074/jbc.M115.697789
Molecular Basis and Consequences of the Cytochrome c-tRNA Interaction
Abstract
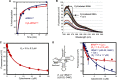
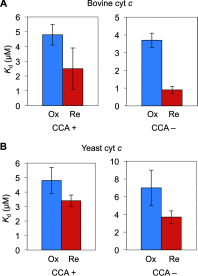
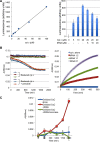
The intrinsic apoptosis pathway occurs through the release of mitochondrial cytochrome c to the cytosol, where it promotes activation of the caspase family of proteases. The observation that tRNA binds to cytochrome c revealed a previously unexpected mode of apoptotic regulation. However, the molecular characteristics of this interaction, and its impact on each interaction partner, are not well understood. Using a novel fluorescence assay, we show here that cytochrome c binds to tRNA with an affinity comparable with other tRNA-protein binding interactions and with a molecular ratio of ∼3:1. Cytochrome c recognizes the tertiary structural features of tRNA, particularly in the core region. This binding is independent of the charging state of tRNA but is regulated by the redox state of cytochrome c. Compared with reduced cytochrome c, oxidized cytochrome c binds to tRNA with a weaker affinity, which correlates with its stronger pro-apoptotic activity. tRNA binding both facilitates cytochrome c reduction and inhibits the peroxidase activity of cytochrome c, which is involved in its release from mitochondria. Together, these findings provide new insights into the cytochrome c-tRNA interaction and apoptotic regulation.
Keywords: apoptosis; caspase; cytochrome c; mitochondrial apoptosis; transfer RNA (tRNA).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Ow Y. P., Green D. R., Hao Z., and Mak T. W. (2008) Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9, 532–542 - PubMed
-
- Liu X., Kim C. N., Yang J., Jemmerson R., and Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 - PubMed
-
- Zou H., Henzel W. J., Liu X., Lutschg A., and Wang X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 - PubMed
-
- Schafer Z. T., and Kornbluth S. (2006) The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell 10, 549–561 - PubMed
-
- Riedl S. J., and Salvesen G. S. (2007) The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

