Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies
- PMID: 26961927
- PMCID: PMC4807699
- DOI: 10.3945/ajcn.115.118216
Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies
Abstract
Mendelian randomization (MR) is an increasingly important tool for appraising causality in observational epidemiology. The technique exploits the principle that genotypes are not generally susceptible to reverse causation bias and confounding, reflecting their fixed nature and Mendel’s first and second laws of inheritance. The approach is, however, subject to important limitations and assumptions that, if unaddressed or compounded by poor study design, can lead to erroneous conclusions. Nevertheless, the advent of 2-sample approaches (in which exposure and outcome are measured in separate samples) and the increasing availability of open-access data from large consortia of genome-wide association studies and population biobanks mean that the approach is likely to become routine practice in evidence synthesis and causal inference research. In this article we provide an overview of the design, analysis, and interpretation of MR studies, with a special emphasis on assumptions and limitations. We also consider different analytic strategies for strengthening causal inference. Although impossible to prove causality with any single approach, MR is a highly cost-effective strategy for prioritizing intervention targets for disease prevention and for strengthening the evidence base for public health policy.
Figures

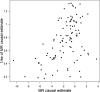
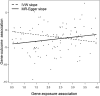
Comment in
-
Health economics and health insurance aspects of Mendelian randomization.Am J Clin Nutr. 2016 Dec;104(6):1720. doi: 10.3945/ajcn.116.137422. Am J Clin Nutr. 2016. PMID: 27935526 No abstract available.
References
-
- Phillips AN, Davey Smith G. How independent are “independent” effects? Relative risk estimation when correlated exposures are measured imprecisely. J Clin Epidemiol 1991;44:1223–31. - PubMed
MeSH terms
Substances
Grants and funding
- G0800270/MRC_/Medical Research Council/United Kingdom
- RG/13/13/30194/BHF_/British Heart Foundation/United Kingdom
- 19169/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_12013/3/MRC_/Medical Research Council/United Kingdom
- RG/08/014/24067/BHF_/British Heart Foundation/United Kingdom
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- MC_UP_1302/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- 20138/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_00002/3/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/2/MRC_/Medical Research Council/United Kingdom
- MC_UU_00002/7/MRC_/Medical Research Council/United Kingdom
- MC_EX_MR/L012286/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_12013/9/MRC_/Medical Research Council/United Kingdom
- MR/N501906/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

