Cross-species functional analyses reveal shared and separate roles for Sox11 in frog primary neurogenesis and mouse cortical neuronal differentiation
- PMID: 26962049
- PMCID: PMC4890661
- DOI: 10.1242/bio.015404
Cross-species functional analyses reveal shared and separate roles for Sox11 in frog primary neurogenesis and mouse cortical neuronal differentiation
Abstract
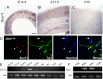
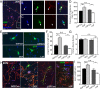
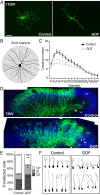
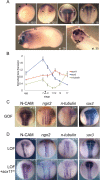
A well-functioning brain requires production of the correct number and types of cells during development; cascades of transcription factors are essential for cellular coordination. Sox proteins are transcription factors that affect various processes in the development of the nervous system. Sox11, a member of the SoxC family, is expressed in differentiated neurons and supports neuronal differentiation in several systems. To understand how generalizable the actions of Sox11 are across phylogeny, its function in the development of the frog nervous system and the mouse cerebral cortex were compared. Expression of Sox11 is largely conserved between these species; in the developing frog, Sox11 is expressed in the neural plate, neural tube and throughout the segmented brain, while in the mouse cerebral cortex, Sox11 is expressed in differentiated zones, including the preplate, subplate, marginal zone and cortical plate. In both frog and mouse, data demonstrate that Sox11 supports a role in promoting neuronal differentiation, with Sox11-positive cells expressing pan-neural markers and becoming morphologically complex. However, frog and mouse Sox11 cannot substitute for one another; a functional difference likely reflected in sequence divergence. Thus, Sox11 appears to act similarly in subserving neuronal differentiation but is species-specific in frog neural development and mouse corticogenesis.
Keywords: Neural development; Neuronal differentiation; Sox transcription factor.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

