An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation
- PMID: 26966090
- PMCID: PMC5043424
- DOI: 10.1182/blood-2016-01-695759
An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation
Abstract
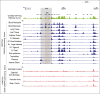
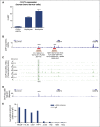
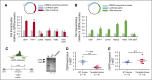
Neutrophilic differentiation is dependent on CCAAT enhancer-binding protein α (C/EBPα), a transcription factor expressed in multiple organs including the bone marrow. Using functional genomic technologies in combination with clustered regularly-interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 genome editing and in vivo mouse modeling, we show that CEBPA is located in a 170-kb topological-associated domain that contains 14 potential enhancers. Of these, 1 enhancer located +42 kb from CEBPA is active and engages with the CEBPA promoter in myeloid cells only. Germ line deletion of the homologous enhancer in mice in vivo reduces Cebpa levels exclusively in hematopoietic stem cells (HSCs) and myeloid-primed progenitor cells leading to severe defects in the granulocytic lineage, without affecting any other Cebpa-expressing organ studied. The enhancer-deleted progenitor cells lose their myeloid transcription program and are blocked in differentiation. Deletion of the enhancer also causes loss of HSC maintenance. We conclude that a single +42-kb enhancer is essential for CEBPA expression in myeloid cells only.
© 2016 by The American Society of Hematology.
Figures




Comment in
-
The potential of a single enhancer.Blood. 2016 Jun 16;127(24):2943-4. doi: 10.1182/blood-2016-03-706218. Blood. 2016. PMID: 27313321 Free PMC article.
References
-
- Miyamoto T, Iwasaki H, Reizis B, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3(1):137–147. - PubMed
-
- Pina C, Fugazza C, Tipping AJ, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14(3):287–294. - PubMed
-
- Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):26–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

