Novel Approach for High-Throughput Metabolic Screening of Whole Plants by Stable Isotopes
- PMID: 26966172
- PMCID: PMC4854671
- DOI: 10.1104/pp.15.01217
Novel Approach for High-Throughput Metabolic Screening of Whole Plants by Stable Isotopes
Abstract
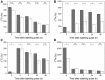
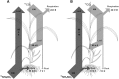
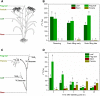
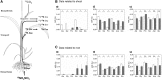
Here, we demonstrate whole-plant metabolic profiling by stable isotope labeling and combustion isotope-ratio mass spectrometry for precise quantification of assimilation, translocation, and molecular reallocation of (13)CO2 and (15)NH4NO3 The technology was applied to rice (Oryza sativa) plants at different growth stages. For adult plants, (13)CO2 labeling revealed enhanced carbon assimilation of the flag leaf from flowering to late grain-filling stage, linked to efficient translocation into the panicle. Simultaneous (13)CO2 and (15)NH4NO3 labeling with hydroponically grown seedlings was used to quantify the relative distribution of carbon and nitrogen. Two hours after labeling, assimilated carbon was mainly retained in the shoot (69%), whereas 7% entered the root and 24% was respired. Nitrogen, taken up via the root, was largely translocated into the shoot (85%). Salt-stressed seedlings showed decreased uptake and translocation of nitrogen (69%), whereas carbon metabolism was unaffected. Coupled to a gas chromatograph, labeling analysis provided enrichment of proteinogenic amino acids. This revealed significant protein synthesis in the panicle of adult plants, whereas protein biosynthesis in adult leaves was 8-fold lower than that in seedling shoots. Generally, amino acid enrichment was similar among biosynthetic families and allowed us to infer labeling dynamics of their precursors. On this basis, early and strong (13)C enrichment of Embden-Meyerhof-Parnas pathway and pentose phosphate pathway intermediates indicated high activity of these routes. Applied to mode-of-action analysis of herbicides, the approach showed severe disturbance in the synthesis of branched-chain amino acids upon treatment with imazapyr. The established technology displays a breakthrough for quantitative high-throughput plant metabolic phenotyping.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Albacete AA, Martínez-Andújar C, Pérez-Alfocea F (2014) Hormonal and metabolic regulation of source-sink relations under salinity and drought: from plant survival to crop yield stability. Biotechnol Adv 32: 12–30 - PubMed
-
- Allen DK, Libourel IGL, Shachar-Hill Y (2009) Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ 32: 1241–1257 - PubMed
-
- Arp WJ. (1991) Effects of source-sink relations on photosynthetic acclimation to elevated CO2. Plant Cell Environ 14: 869–875
-
- Cegelski L, Schaefer J (2005) Glycine metabolism in intact leaves by in vivo 13C and 15N labeling. J Biol Chem 280: 39238–39245 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

