Molecular Occupancy of Nanodot Arrays
- PMID: 26966946
- PMCID: PMC5337305
- DOI: 10.1021/acsnano.5b07425
Molecular Occupancy of Nanodot Arrays
Abstract
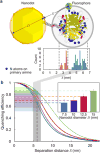
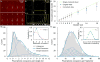
Single-molecule nanodot arrays, in which a biomolecule of choice (protein, nucleic acid, etc.) is bound to a metallic nanoparticle on a solid substrate, are becoming an increasingly important tool in the study of biomolecular and cellular interactions. We have developed an on-chip measurement protocol to monitor and control the molecular occupancy of nanodots. Arrays of widely spaced nanodots and nanodot clusters were fabricated on glass surfaces by nanolithography and functionalized with fluorescently labeled proteins. The molecular occupancy was determined by monitoring individual fluorophore bleaching events, while accounting for fluorescence quenching effects. We found that the occupancy can be interpreted as a packing problem, and depends on nanodot size and binding ligand concentration, where the latter is easily adjusted to compensate the flexibility of dimension control in nanofabrication. The results are scalable with nanodot cluster size, extending to large area close packed arrays. As an example, the nanoarray platform was used to probe the geometric requirement of T-cell activation at the single-molecule level.
Keywords: electron beam lithography; fluorescence quenching; molecular occupancy; photobleaching; single-molecule assays; stoichiometry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen LS, Chernikoff D;, et al. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science. 2010;327:78–81. - PubMed
-
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133–138. - PubMed
-
- Lynch M, Mosher C, Huff J, Nettikadan S, Johnson J, Henderson E. Functional Protein Nanoarrays for Biomarker Profiling. Proteomics. 2004;4:1695–1702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

