The 3D Genome as Moderator of Chromosomal Communication
- PMID: 26967279
- PMCID: PMC4788811
- DOI: 10.1016/j.cell.2016.02.007
The 3D Genome as Moderator of Chromosomal Communication
Abstract
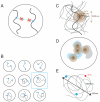
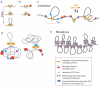
Proper expression of genes requires communication with their regulatory elements that can be located elsewhere along the chromosome. The physics of chromatin fibers imposes a range of constraints on such communication. The molecular and biophysical mechanisms by which chromosomal communication is established, or prevented, have become a topic of intense study, and important roles for the spatial organization of chromosomes are being discovered. Here we present a view of the interphase 3D genome characterized by extensive physical compartmentalization and insulation on the one hand and facilitated long-range interactions on the other. We propose the existence of topological machines dedicated to set up and to exploit a 3D genome organization to both promote and censor communication along and between chromosomes.
Keywords: CTCF; Hi-C; biophysics; cohesin; condensin; domains; enhancer; gene expression; gene regulation; polymers; promoter; simulations.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. - PubMed
-
- Barakat TS, Loos F, van Staveren S, Myronova E, Ghazvini M, Grootegoed JA, Gribnau J. The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of X-pairing. Mol Cell. 2014;53:965–978. - PubMed
-
- Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 R01 AI 117839/AI/NIAID NIH HHS/United States
- R01 GM112720/GM/NIGMS NIH HHS/United States
- R01 GM114190/GM/NIGMS NIH HHS/United States
- U01 DA 040588/DA/NIDA NIH HHS/United States
- R01 HG003143/HG/NHGRI NIH HHS/United States
- R01 GM 112720/GM/NIGMS NIH HHS/United States
- U01 HG007910/HG/NHGRI NIH HHS/United States
- U54 DK107980/DK/NIDDK NIH HHS/United States
- R01 AI117839/AI/NIAID NIH HHS/United States
- Howard Hughes Medical Institute/United States
- U01 DA040588/DA/NIDA NIH HHS/United States
- U54 HG007010/HG/NHGRI NIH HHS/United States
- U54 CA193419/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

