Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs
- PMID: 26968199
- PMCID: PMC4817775
- DOI: 10.1101/gr.199703.115
Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs
Abstract
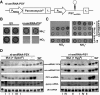
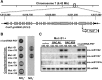
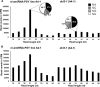
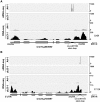
We describe here a forward genetic screen to investigate the biogenesis, mode of action, and biological function of miRNA-mediated RNA silencing in the model algal species,Chlamydomonas reinhardtii Among the mutants from this screen, there were three at Dicer-like 3 that failed to produce both miRNAs and siRNAs and others affecting diverse post-biogenesis stages of miRNA-mediated silencing. The DCL3-dependent siRNAs fell into several classes including transposon- and repeat-derived siRNAs as in higher plants. The DCL3-dependent miRNAs differ from those of higher plants, however, in that many of them are derived from mRNAs or from the introns of pre-mRNAs. Transcriptome analysis of the wild-type and dcl3 mutant strains revealed a further difference from higher plants in that the sRNAs are rarely negative switches of mRNA accumulation. The few transcripts that were more abundant in dcl3 mutant strains than in wild-type cells were not due to sRNA-targeted RNA degradation but to direct DCL3 cleavage of miRNA and siRNA precursor structures embedded in the untranslated (and translated) regions of the mRNAs. Our analysis reveals that the miRNA-mediated RNA silencing in C. reinhardtii differs from that of higher plants and informs about the evolution and function of this pathway in eukaryotes.
© 2016 Valli et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Baulcombe D. 2004. RNA silencing in plants. Nature 431: 356–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
