Does Gene Tree Discordance Explain the Mismatch between Macroevolutionary Models and Empirical Patterns of Tree Shape and Branching Times?
- PMID: 26968785
- PMCID: PMC4911941
- DOI: 10.1093/sysbio/syw019
Does Gene Tree Discordance Explain the Mismatch between Macroevolutionary Models and Empirical Patterns of Tree Shape and Branching Times?
Abstract
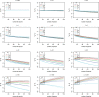
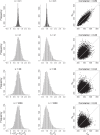
Classic null models for speciation and extinction give rise to phylogenies that differ in distribution from empirical phylogenies. In particular, empirical phylogenies are less balanced and have branching times closer to the root compared to phylogenies predicted by common null models. This difference might be due to null models of the speciation and extinction process being too simplistic, or due to the empirical datasets not being representative of random phylogenies. A third possibility arises because phylogenetic reconstruction methods often infer gene trees rather than species trees, producing an incongruity between models that predict species tree patterns and empirical analyses that consider gene trees. We investigate the extent to which the difference between gene trees and species trees under a combined birth-death and multispecies coalescent model can explain the difference in empirical trees and birth-death species trees. We simulate gene trees embedded in simulated species trees and investigate their difference with respect to tree balance and branching times. We observe that the gene trees are less balanced and typically have branching times closer to the root than the species trees. Empirical trees from TreeBase are also less balanced than our simulated species trees, and model gene trees can explain an imbalance increase of up to 8% compared to species trees. However, we see a much larger imbalance increase in empirical trees, about 100%, meaning that additional features must also be causing imbalance in empirical trees. This simulation study highlights the necessity of revisiting the assumptions made in phylogenetic analyses, as these assumptions, such as equating the gene tree with the species tree, might lead to a biased conclusion.
Keywords: Birth–death process; genealogy; multispecies coalescent; phylogeny.
© The Author(s) 2016. Published by Oxford University Press, on behalf of the Society of Systematic Biologists.
Figures


Similar articles
-
Age-dependent speciation can explain the shape of empirical phylogenies.Syst Biol. 2015 May;64(3):432-40. doi: 10.1093/sysbio/syv001. Epub 2015 Jan 8. Syst Biol. 2015. PMID: 25575504 Free PMC article.
-
How Ecology and Landscape Dynamics Shape Phylogenetic Trees.Syst Biol. 2015 Jul;64(4):590-607. doi: 10.1093/sysbio/syv014. Epub 2015 Mar 13. Syst Biol. 2015. PMID: 25771083
-
What is the danger of the anomaly zone for empirical phylogenetics?Syst Biol. 2009 Oct;58(5):527-36. doi: 10.1093/sysbio/syp047. Epub 2009 Aug 26. Syst Biol. 2009. PMID: 20525606
-
The inference of gene trees with species trees.Syst Biol. 2015 Jan;64(1):e42-62. doi: 10.1093/sysbio/syu048. Epub 2014 Jul 28. Syst Biol. 2015. PMID: 25070970 Free PMC article. Review.
-
Coalescent methods for estimating phylogenetic trees.Mol Phylogenet Evol. 2009 Oct;53(1):320-8. doi: 10.1016/j.ympev.2009.05.033. Epub 2009 Jun 6. Mol Phylogenet Evol. 2009. PMID: 19501178 Review.
Cited by
-
Anomalous networks under the multispecies coalescent: theory and prevalence.J Math Biol. 2024 Feb 19;88(3):29. doi: 10.1007/s00285-024-02050-7. J Math Biol. 2024. PMID: 38372830
-
Probabilities of Unranked and Ranked Anomaly Zones under Birth-Death Models.Mol Biol Evol. 2020 May 1;37(5):1480-1494. doi: 10.1093/molbev/msz305. Mol Biol Evol. 2020. PMID: 31860090 Free PMC article.
-
An analytical upper bound on the number of loci required for all splits of a species tree to appear in a set of gene trees.BMC Bioinformatics. 2016 Nov 11;17(Suppl 14):417. doi: 10.1186/s12859-016-1266-4. BMC Bioinformatics. 2016. PMID: 28185570 Free PMC article.
-
Taxonomic Uncertainty and the Anomaly Zone: Phylogenomics Disentangle a Rapid Radiation to Resolve Contentious Species (Gila robusta Complex) in the Colorado River.Genome Biol Evol. 2021 Sep 1;13(9):evab200. doi: 10.1093/gbe/evab200. Genome Biol Evol. 2021. PMID: 34432005 Free PMC article.
-
Comprehensive Phylogenetic Analysis of Bovine Non-aureus Staphylococci Species Based on Whole-Genome Sequencing.Front Microbiol. 2016 Dec 20;7:1990. doi: 10.3389/fmicb.2016.01990. eCollection 2016. Front Microbiol. 2016. PMID: 28066335 Free PMC article.
References
-
- Agapow P.M., Purvis A. 2002. Power of eight tree shape statistics to detect nonrandom diversification: a comparison by simulation of two models of cladogenesis. Syst. Biol. 51: 866–872. - PubMed
-
- Aldous D., Pemantle R., editors. 1996. Random discrete structures, vol. 76 of The IMA volumes in mathematics and its applications. Springer, New York; p. 1–18.
-
- Aldous D., Popovic L. 2005. A critical branching process model for biodiversity. Adv. Appl. Prob. 37: 1094–1115.
-
- Aldous D.J. 2001. Stochastic models and descriptive statistics for phylogenetic trees, from Yule to today. Statist. Sci. 16: 23–34.
-
- Blum M.G.B., François O. 2006. Which random processes describe the tree of life? A large-scale study of phylogenetic tree imbalance. Syst. Biol. 55: 685–691. - PubMed
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

