Causes of molecular convergence and parallelism in protein evolution
- PMID: 26972590
- PMCID: PMC5482790
- DOI: 10.1038/nrg.2016.11
Causes of molecular convergence and parallelism in protein evolution
Abstract
To what extent is the convergent evolution of protein function attributable to convergent or parallel changes at the amino acid level? The mutations that contribute to adaptive protein evolution may represent a biased subset of all possible beneficial mutations owing to mutation bias and/or variation in the magnitude of deleterious pleiotropy. A key finding is that the fitness effects of amino acid mutations are often conditional on genetic background. This context dependence (epistasis) can reduce the probability of convergence and parallelism because it reduces the number of possible mutations that are unconditionally acceptable in divergent genetic backgrounds. Here, I review factors that influence the probability of replicated evolution at the molecular level.
Conflict of interest statement
The author declares no competing interests.
Figures
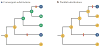


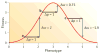
References
-
- Wake DB. Homoplasy: the result of natural selection, or evidence of design limitations? Am Naturalist. 1991;138:543–567.
-
- Losos JB. Covergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. - PubMed
-
- Chevin LM, Martin G, Lenormand T. Fisher’s model and the genomics of adaptation: restricted pleiotropy, heterogeneous mutation, and parallel evolution. Evolution. 2010;64:3213–3231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

