IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis
- PMID: 26972847
- PMCID: PMC4793079
- DOI: 10.1038/ncomms10791
IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis
Abstract
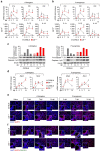
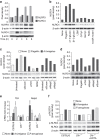
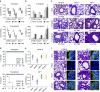
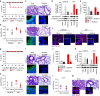
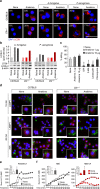
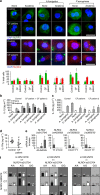
Dysregulated inflammasome activation contributes to respiratory infections and pathologic airway inflammation. Through basic and translational approaches involving murine models and human genetic epidemiology, we show here the importance of the different inflammasomes in regulating inflammatory responses in mice and humans with cystic fibrosis (CF), a life-threatening disorder of the lungs and digestive system. While both contributing to pathogen clearance, NLRP3 more than NLRC4 contributes to deleterious inflammatory responses in CF and correlates with defective NLRC4-dependent IL-1Ra production. Disease susceptibility in mice and microbial colonization in humans occurs in conditions of genetic deficiency of NLRC4 or IL-1Ra and can be rescued by administration of the recombinant IL-1Ra, anakinra. These results indicate that pathogenic NLRP3 activity in CF could be negatively regulated by IL-1Ra and provide a proof-of-concept evidence that inflammasomes are potential targets to limit the pathological consequences of microbial colonization in CF.
Figures


References
-
- Corvol H. et al. Distinct cytokine production by lung and blood neutrophils from children with cystic fibrosis. Am. J. Physiol. Lung. Cell Mol. Physiol. 284, L997–L1003 (2003). - PubMed
-
- Iannitti R. G. et al. Th17/Treg imbalance in murine cystic fibrosis is linked to indoleamine 2,3-dioxygenase deficiency but corrected by kynurenines. Am. J. Respir. Crit. Care Med. 187, 609–620 (2013). - PubMed
-
- Robinson K. M. & Alcorn J. F. T-cell immunotherapy in cystic fibrosis: weighing the risk/reward. Am. J. Respir. Crit. Care Med. 187, 564–566 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

