Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice
- PMID: 26973250
- PMCID: PMC5057043
- DOI: 10.1038/onc.2016.58
Olfactomedin 4 deletion induces colon adenocarcinoma in ApcMin/+ mice
Abstract
Colon carcinogenesis is a multiple-step process involving the accumulation of a series of genetic and epigenetic alterations. The most commonly initiating event of intestinal carcinogenesis is mutation of the adenomatous polyposis coli (APC) gene, which leads to activation of the Wnt/β-catenin pathway. Olfactomedin 4 (OLFM4) has emerged as an intestinal stem-cell marker, but its biological function in the intestine remains to be determined. Here we show that Olfm4 deletion induced colon adenocarcinoma in the distal colon of ApcMin/+ mice. Mechanistically, we found that OLFM4 is a target gene of the Wnt/β-catenin pathway and can downregulate β-catenin signaling by competing with Wnt ligands for binding to Frizzled receptors, as well as by inhibition of the Akt-GSK-3β (Akt-glycogen synthase kinase-3β) pathway. We have shown that both Wnt and nuclear factor-κB (NF-κB) signaling were boosted in tumor tissues of Apc Olfm4 double-mutant mice. These data establish OLFM4 as a critical negative regulator of the Wnt/β-catenin and NF-κB pathways that inhibits colon-cancer development initiated by APC mutation. In addition, Olfm4 deletion significantly enhanced intestinal-crypt proliferation and inflammation induced by azoxymethane/dextran sodium sulfate. Thus, OLFM4 has an important role in the regulation of intestinal inflammation and tumorigenesis, and could be a potential therapeutic target for intestinal malignant tumors. Unlike the human colonic epithelium, the mouse colonic epithelium does not express OLFM4, but nevertheless, systemic OLFM4 deletion promotes colon tumorigenesis and that loss from mucosal neutrophils may have a role to play.
Figures

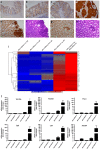


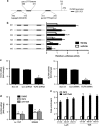
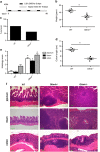
References
-
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol 2011; 6: 479–507. - PubMed
-
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell 2000; 103: 311–320. - PubMed
-
- Inomata M, Ochiai A, Akimoto S, Kitano S, Hirohashi S. Alteration of beta- catenin expression in colonic epithelial cells of familial adenomatous polyposis patients. Cancer Res 1996; 56: 2213–2217. - PubMed
-
- Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 2005; 5: 749–759. - PubMed
-
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013; 152: 25–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

