Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses
- PMID: 26973327
- PMCID: PMC4989048
- DOI: 10.1016/j.joca.2016.03.007
Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses
Abstract
Objectives: Joint degeneration in osteoarthritis (OA) is characterised by damage and loss of articular cartilage. The pattern of loss is consistent with damage occurring only where the mechanical loading is high. We have investigated using RNA-sequencing (RNA-seq) and systems analyses the changes that occur in damaged OA cartilage by comparing it with intact cartilage from the same joint.
Methods: Cartilage was obtained from eight OA patients undergoing total knee replacement. RNA was extracted from cartilage on the damaged distal medial condyle (DMC) and the intact posterior lateral condyle (PLC). RNA-seq was performed to identify differentially expressed genes (DEGs) and systems analyses applied to identify dysregulated pathways.
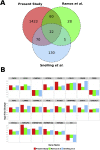
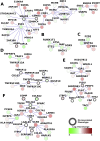
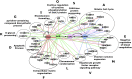
Results: In the damaged OA cartilage, there was decreased expression of chondrogenic genes SOX9, SOX6, COL11A2, COL9A1/2/3, ACAN and HAPLN1; increases in non-chondrogenic genes COL1A1, COMP and FN1; an altered pattern of secreted proteinase expression; but no expression of major inflammatory cytokines. Systems analyses by PhenomeExpress revealed significant sub-networks of DEGs including mitotic cell cycle, Wnt signalling, apoptosis and matrix organisation that were influenced by a core of altered transcription factors (TFs), FOSL1, AHR, E2F1 and FOXM1.
Conclusions: Gene expression changes in damaged cartilage suggested a signature non-chondrogenic response of altered matrix protein and secreted proteinase expression. There was evidence of a damage response in this late OA cartilage, which surprisingly showed features detected experimentally in the early response of cartilage to mechanical overload. PhenomeExpress analysis identified a hub of DEGs linked by a core of four differentially regulated TFs.
Keywords: Cartilage; Osteoarthritis; PhenomeExpress; RNA-seq; Systems biology.
Copyright © 2016 Osteoarthritis Research Society International. Published by Elsevier Ltd. All rights reserved.
Figures

References
-
- Goldring M.B. Update on the biology of the chondrocyte and new approaches to treating cartilage diseases. Best Pract Res Clin Rheumatol. 2006;20:1003–1025. - PubMed
-
- Aigner T., Fundel K., Saas J., Gebhard P.M., Haag J., Weiss T. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

