Organohalide Respiring Bacteria and Reductive Dehalogenases: Key Tools in Organohalide Bioremediation
- PMID: 26973626
- PMCID: PMC4771760
- DOI: 10.3389/fmicb.2016.00249
Organohalide Respiring Bacteria and Reductive Dehalogenases: Key Tools in Organohalide Bioremediation
Abstract
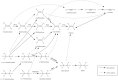
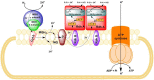
Organohalides are recalcitrant pollutants that have been responsible for substantial contamination of soils and groundwater. Organohalide-respiring bacteria (ORB) provide a potential solution to remediate contaminated sites, through their ability to use organohalides as terminal electron acceptors to yield energy for growth (i.e., organohalide respiration). Ideally, this process results in non- or lesser-halogenated compounds that are mostly less toxic to the environment or more easily degraded. At the heart of these processes are reductive dehalogenases (RDases), which are membrane bound enzymes coupled with other components that facilitate dehalogenation of organohalides to generate cellular energy. This review focuses on RDases, concentrating on those which have been purified (partially or wholly) and functionally characterized. Further, the paper reviews the major bacteria involved in organohalide breakdown and the evidence for microbial evolution of RDases. Finally, the capacity for using ORB in a bioremediation and bioaugmentation capacity are discussed.
Keywords: Dehalobacter; Dehalococcoides; bioremediation; organohalide respiration; reductive dehalogenase.
Figures

References
-
- Boyer A., Pagé-BéLanger R., Saucier M., Villemur R., Lépine F., Juteau P., et al. (2003). Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6-trichlorophenol from Desulfitobacterium frappieri PCP-1. Biochem. J. 373, 297–303. 10.1042/bj20021837 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

