Bacterial antisense RNAs are mainly the product of transcriptional noise
- PMID: 26973873
- PMCID: PMC4783119
- DOI: 10.1126/sciadv.1501363
Bacterial antisense RNAs are mainly the product of transcriptional noise
Abstract
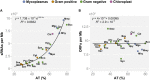
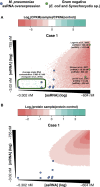
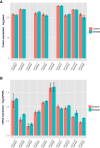
cis-Encoded antisense RNAs (asRNAs) are widespread along bacterial transcriptomes. However, the role of most of these RNAs remains unknown, and there is an ongoing discussion as to what extent these transcripts are the result of transcriptional noise. We show, by comparative transcriptomics of 20 bacterial species and one chloroplast, that the number of asRNAs is exponentially dependent on the genomic AT content and that expression of asRNA at low levels exerts little impact in terms of energy consumption. A transcription model simulating mRNA and asRNA production indicates that the asRNA regulatory effect is only observed above certain expression thresholds, substantially higher than physiological transcript levels. These predictions were verified experimentally by overexpressing nine different asRNAs in Mycoplasma pneumoniae. Our results suggest that most of the antisense transcripts found in bacteria are the consequence of transcriptional noise, arising at spurious promoters throughout the genome.
Keywords: RNA; bacterial antisense RNAs.
Figures
References
-
- Altuvia S., Identification of bacterial small non-coding RNAs: Experimental approaches. Curr. Opin. Microbiol. 10, 257–261 (2007). - PubMed
-
- Wade J. T., Grainger D. C., Pervasive transcription: Illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

