Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype
- PMID: 26973881
- PMCID: PMC4784261
- DOI: 10.1172/jci.insight.85841
Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype
Abstract
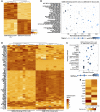
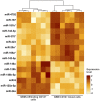
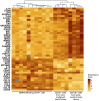
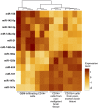
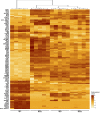
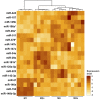
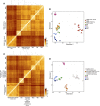
Glioblastomas are highly infiltrated by diverse immune cells, including microglia, macrophages, and myeloid-derived suppressor cells (MDSCs). Understanding the mechanisms by which glioblastoma-associated myeloid cells (GAMs) undergo metamorphosis into tumor-supportive cells, characterizing the heterogeneity of immune cell phenotypes within glioblastoma subtypes, and discovering new targets can help the design of new efficient immunotherapies. In this study, we performed a comprehensive battery of immune phenotyping, whole-genome microarray analysis, and microRNA expression profiling of GAMs with matched blood monocytes, healthy donor monocytes, normal brain microglia, nonpolarized M0 macrophages, and polarized M1, M2a, M2c macrophages. Glioblastoma patients had an elevated number of monocytes relative to healthy donors. Among CD11b+ cells, microglia and MDSCs constituted a higher percentage of GAMs than did macrophages. GAM profiling using flow cytometry studies revealed a continuum between the M1- and M2-like phenotype. Contrary to current dogma, GAMs exhibited distinct immunological functions, with the former aligned close to nonpolarized M0 macrophages.
Figures



References
-
- Badie B, Schartner JM. Flow cytometric characterization of tumor-associated macrophages in experimental gliomas. Neurosurgery. 2000;46(4):957–961. - PubMed
-
- Wierzba-Bobrowicz T, Kuchna I, Matyja E. Reaction of microglial cells in human astrocytomas (preliminary report) Folia Neuropathol. 1994;32(4):251–252. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

