Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer
- PMID: 26975198
- PMCID: PMC4791797
- DOI: 10.1186/s13058-016-0690-8
Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer
Abstract
Background: Triple-negative breast cancer (TNBC) is a highly heterogeneous group of cancers, and molecular subtyping is necessary to better identify molecular-based therapies. While some classifiers have been established, no one has integrated the expression profiles of long noncoding RNAs (lncRNAs) into such subtyping criterions. Considering the emerging important role of lncRNAs in cellular processes, a novel classification integrating transcriptome profiles of both messenger RNA (mRNA) and lncRNA would help us better understand the heterogeneity of TNBC.
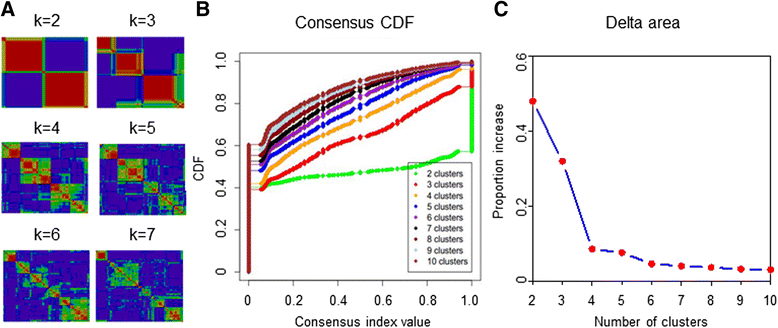
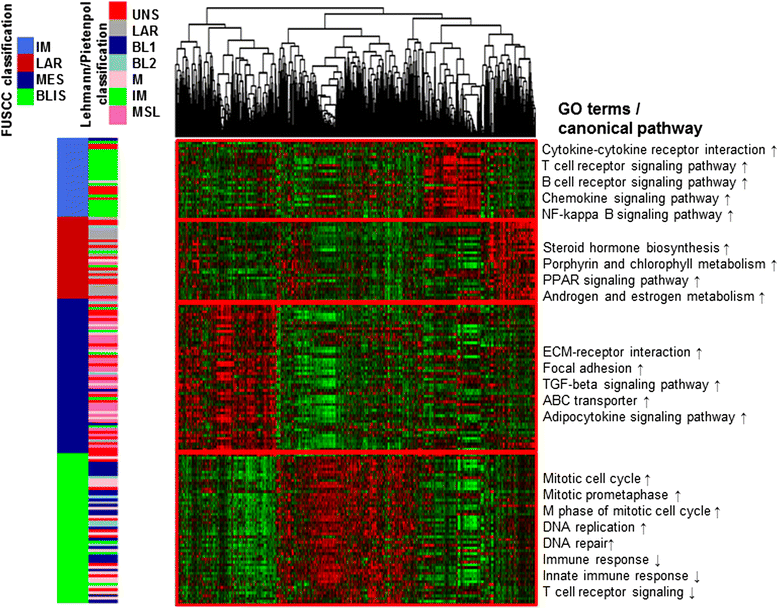
Methods: Using human transcriptome microarrays, we analyzed the transcriptome profiles of 165 TNBC samples. We used k-means clustering and empirical cumulative distribution function to determine optimal number of TNBC subtypes. Gene Ontology (GO) and pathway analyses were applied to determine the main function of the subtype-specific genes and pathways. We conducted co-expression network analyses to identify interactions between mRNAs and lncRNAs.
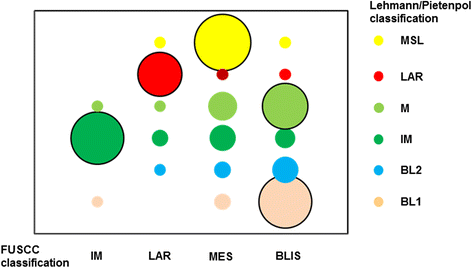
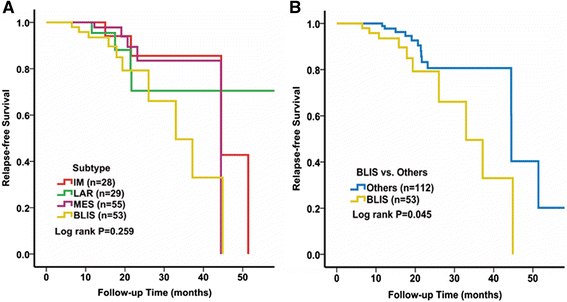
Results: All of the 165 TNBC tumors were classified into four distinct clusters, including an immunomodulatory subtype (IM), a luminal androgen receptor subtype (LAR), a mesenchymal-like subtype (MES) and a basal-like and immune suppressed (BLIS) subtype. The IM subtype had high expressions of immune cell signaling and cytokine signaling genes. The LAR subtype was characterized by androgen receptor signaling. The MES subtype was enriched with growth factor signaling pathways. The BLIS subtype was characterized by down-regulation of immune response genes, activation of cell cycle, and DNA repair. Patients in this subtype experienced worse recurrence-free survival than others (log rank test, P = 0.045). Subtype-specific lncRNAs were identified, and their possible biological functions were predicted using co-expression network analyses.
Conclusions: We developed a novel TNBC classification system integrating the expression profiles of both mRNAs and lncRNAs and determined subtype-specific lncRNAs that are potential biomarkers and targets. If further validated in a larger population, our novel classification system could facilitate patient counseling and individualize treatment of TNBC.
Keywords: Long non-coding RNA; Messenger RNA; Molecular subtypes; Transcriptome analysis; Triple-negative breast cancer.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

