Identification of tissue-specific cell death using methylation patterns of circulating DNA
- PMID: 26976580
- PMCID: PMC4822610
- DOI: 10.1073/pnas.1519286113
Identification of tissue-specific cell death using methylation patterns of circulating DNA
Abstract
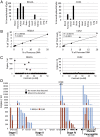
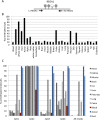
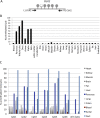
Minimally invasive detection of cell death could prove an invaluable resource in many physiologic and pathologic situations. Cell-free circulating DNA (cfDNA) released from dying cells is emerging as a diagnostic tool for monitoring cancer dynamics and graft failure. However, existing methods rely on differences in DNA sequences in source tissues, so that cell death cannot be identified in tissues with a normal genome. We developed a method of detecting tissue-specific cell death in humans based on tissue-specific methylation patterns in cfDNA. We interrogated tissue-specific methylome databases to identify cell type-specific DNA methylation signatures and developed a method to detect these signatures in mixed DNA samples. We isolated cfDNA from plasma or serum of donors, treated the cfDNA with bisulfite, PCR-amplified the cfDNA, and sequenced it to quantify cfDNA carrying the methylation markers of the cell type of interest. Pancreatic β-cell DNA was identified in the circulation of patients with recently diagnosed type-1 diabetes and islet-graft recipients; oligodendrocyte DNA was identified in patients with relapsing multiple sclerosis; neuronal/glial DNA was identified in patients after traumatic brain injury or cardiac arrest; and exocrine pancreas DNA was identified in patients with pancreatic cancer or pancreatitis. This proof-of-concept study demonstrates that the tissue origins of cfDNA and thus the rate of death of specific cell types can be determined in humans. The approach can be adapted to identify cfDNA derived from any cell type in the body, offering a minimally invasive window for diagnosing and monitoring a broad spectrum of human pathologies as well as providing a better understanding of normal tissue dynamics.
Keywords: circulating DNA; diagnosis; methylation.
Conflict of interest statement
Conflict of interest statement: C.D. and M.G. are the inventors of antibodies directed against human pancreatic cells, HPx1/HIC0-3B3 and HPd3/DHIC5-4D9. Oregon Health & Science University (OHSU) has commercially licensed this technology. This potential conflict of interest has been reviewed and managed by OHSU.
Figures









References
-
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. - PubMed
-
- Bianchi DW, et al. CARE Study Group DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. - PubMed
-
- Lo YM, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

