LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior
- PMID: 26976584
- PMCID: PMC4822633
- DOI: 10.1073/pnas.1511974113
LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior
Abstract
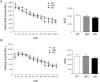
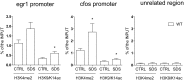
Behavioral changes in response to stressful stimuli can be controlled via adaptive epigenetic changes in neuronal gene expression. Here we indicate a role for the transcriptional corepressor Lysine-Specific Demethylase 1 (LSD1) and its dominant-negative splicing isoform neuroLSD1, in the modulation of emotional behavior. In mouse hippocampus, we show that LSD1 and neuroLSD1 can interact with transcription factor serum response factor (SRF) and set the chromatin state of SRF-targeted genes early growth response 1 (egr1) and c-fos Deletion or reduction of neuro LSD1 in mutant mice translates into decreased levels of activating histone marks at egr1 and c-fos promoters, dampening their psychosocial stress-induced transcription and resulting in low anxiety-like behavior. Administration of suberoylanilide hydroxamine to neuroLSD1(KO)mice reactivates egr1 and c-fos transcription and restores the behavioral phenotype. These findings indicate that LSD1 is a molecular transducer of stressful stimuli as well as a stress-response modifier. Indeed, LSD1 expression itself is increased acutely at both the transcriptional and splicing levels by psychosocial stress, suggesting that LSD1 is involved in the adaptive response to stress.
Keywords: LSD1; SRF; epigenetics; immediate early genes; stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures














References
-
- Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–525. - PubMed
-
- Chattarji S, Tomar A, Suvrathan A, Ghosh S, Rahman MM. Neighborhood matters: divergent patterns of stress-induced plasticity across the brain. Nat Neurosci. 2015;18(10):1364–1375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

