Protein networks identify novel symbiogenetic genes resulting from plastid endosymbiosis
- PMID: 26976593
- PMCID: PMC4822624
- DOI: 10.1073/pnas.1517551113
Protein networks identify novel symbiogenetic genes resulting from plastid endosymbiosis
Abstract
The integration of foreign genetic information is central to the evolution of eukaryotes, as has been demonstrated for the origin of the Calvin cycle and of the heme and carotenoid biosynthesis pathways in algae and plants. For photosynthetic lineages, this coordination involved three genomes of divergent phylogenetic origins (the nucleus, plastid, and mitochondrion). Major hurdles overcome by the ancestor of these lineages were harnessing the oxygen-evolving organelle, optimizing the use of light, and stabilizing the partnership between the plastid endosymbiont and host through retargeting of proteins to the nascent organelle. Here we used protein similarity networks that can disentangle reticulate gene histories to explore how these significant challenges were met. We discovered a previously hidden component of algal and plant nuclear genomes that originated from the plastid endosymbiont: symbiogenetic genes (S genes). These composite proteins, exclusive to photosynthetic eukaryotes, encode a cyanobacterium-derived domain fused to one of cyanobacterial or another prokaryotic origin and have emerged multiple, independent times during evolution. Transcriptome data demonstrate the existence and expression of S genes across a wide swath of algae and plants, and functional data indicate their involvement in tolerance to oxidative stress, phototropism, and adaptation to nitrogen limitation. Our research demonstrates the "recycling" of genetic information by photosynthetic eukaryotes to generate novel composite genes, many of which function in plastid maintenance.
Keywords: endosymbiosis; eukaryote evolution; gene fusion; novel gene origin; photosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

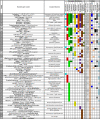

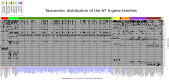

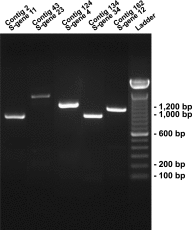

References
-
- Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393(6681):162–165. - PubMed
-
- Reyes-Prieto A, Hackett JD, Soares MB, Bonaldo MF, Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr Biol. 2006;16(23):2320–2325. - PubMed
-
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21(5):809–818. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

