Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis
- PMID: 26976605
- PMCID: PMC4822641
- DOI: 10.1073/pnas.1602532113
Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis
Abstract
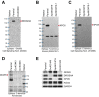
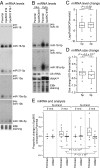
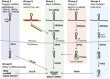
Biogenesis of canonical microRNAs (miRNAs) involves multiple steps: nuclear processing of primary miRNA (pri-miRNA) by DROSHA, nuclear export of precursor miRNA (pre-miRNA) by Export in 5 (XPO5), and cytoplasmic processing of pre-miRNA by DICER. To gain a deeper understanding of the contribution of each of these maturation steps, we deleted DROSHA, XPO5, and DICER in the same human cell line, and analyzed their effects on miRNA biogenesis. Canonical miRNA production was completely abolished in DROSHA-deleted cells, whereas we detected a few DROSHA-independent miRNAs including three previously unidentified noncanonical miRNAs (miR-7706, miR-3615, and miR-1254). In contrast to DROSHA knockout, many canonical miRNAs were still detected without DICER albeit at markedly reduced levels. In the absence of DICER, pre-miRNAs are loaded directly onto AGO and trimmed at the 3' end, yielding miRNAs from the 5' strand (5p miRNAs). Interestingly, in XPO5 knockout cells, most miRNAs are affected only modestly, suggesting that XPO5 is necessary but not critical for miRNA maturation. Our study demonstrates an essential role of DROSHA and an important contribution of DICER in the canonical miRNA pathway, and reveals that the function of XPO5 can be complemented by alternative mechanisms. Thus, this study allows us to understand differential contributions of key biogenesis factors, and provides with valuable resources for miRNA research.
Keywords: DICER; DROSHA; Exportin 5; knockout; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. - PubMed
-
- Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. - PubMed
-
- Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

