Stabilization of a prokaryotic LAT transporter by random mutagenesis
- PMID: 26976827
- PMCID: PMC4810068
- DOI: 10.1085/jgp.201511510
Stabilization of a prokaryotic LAT transporter by random mutagenesis
Abstract
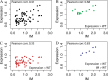
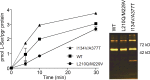
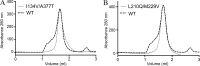
The knowledge of three-dimensional structures at atomic resolution of membrane transport proteins has improved considerably our understanding of their physiological roles and pathological implications. However, most structural biology techniques require an optimal candidate within a protein family for structural determination with (a) reasonable production in heterologous hosts and (b) good stability in detergent micelles. SteT, the Bacillus subtilis L-serine/L-threonine exchanger is the best-known prokaryotic paradigm of the mammalian L-amino acid transporter (LAT) family. Unfortunately, SteT's lousy stability after extracting from the membrane prevents its structural characterization. Here, we have used an approach based on random mutagenesis to engineer stability in SteT. Using a split GFP complementation assay as reporter of protein expression and membrane insertion, we created a library of 70 SteT mutants each containing random replacements of one or two residues situated in the transmembrane domains. Analysis of expression and monodispersity in detergent of this library permitted the identification of evolved versions of SteT with a significant increase in both expression yield and stability in detergent with respect to wild type. In addition, these experiments revealed a correlation between the yield of expression and the stability in detergent micelles. Finally, and based on protein delipidation and relipidation assays together with transport experiments, possible mechanisms of SteT stabilization are discussed. Besides optimizing a member of the LAT family for structural determination, our work proposes a new approach that can be used to optimize any membrane protein of interest.
© 2016 Rodríguez-Banqueri et al.
Figures








Similar articles
-
Split GFP Complementation as Reporter of Membrane Protein Expression and Stability in E. coli: A Tool to Engineer Stability in a LAT Transporter.Methods Mol Biol. 2017;1586:181-195. doi: 10.1007/978-1-4939-6887-9_11. Methods Mol Biol. 2017. PMID: 28470605
-
Role of transmembrane domain 8 in substrate selectivity and translocation of SteT, a member of the L-amino acid transporter (LAT) family.J Biol Chem. 2010 Sep 10;285(37):28764-76. doi: 10.1074/jbc.M110.116632. Epub 2010 Jul 7. J Biol Chem. 2010. PMID: 20610400 Free PMC article.
-
Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: a model for APC transporters.J Biol Chem. 2007 May 4;282(18):13270-81. doi: 10.1074/jbc.M610695200. Epub 2007 Mar 6. J Biol Chem. 2007. PMID: 17344220
-
What is an antidepressant binding site doing in a bacterial transporter?ACS Chem Biol. 2007 Sep 21;2(9):606-9. doi: 10.1021/cb7001818. ACS Chem Biol. 2007. PMID: 17894444 Review.
-
The amino acid transport system b(o,+) and cystinuria.Mol Membr Biol. 2001 Jan-Mar;18(1):21-6. Mol Membr Biol. 2001. PMID: 11396607 Review.
Cited by
-
Directed Evolution of Soluble α-1,2-Fucosyltransferase Using Kanamycin Resistance Protein as a Phenotypic Reporter for Efficient Production of 2'-Fucosyllactose.J Microbiol Biotechnol. 2022 Nov 28;32(11):1471-1478. doi: 10.4014/jmb.2209.09018. Epub 2022 Oct 17. J Microbiol Biotechnol. 2022. PMID: 36437520 Free PMC article.
-
Functional characterization of the alanine-serine-cysteine exchanger of Carnobacterium sp AT7.J Gen Physiol. 2019 Apr 1;151(4):505-517. doi: 10.1085/jgp.201812195. Epub 2019 Jan 29. J Gen Physiol. 2019. PMID: 30696726 Free PMC article.
-
Precise Engineering and Efficient Biosynthesis of Robust and High-Activity Human Haemoglobin for Artificial Oxygen Carriers.Microb Biotechnol. 2025 Mar;18(3):e70128. doi: 10.1111/1751-7915.70128. Microb Biotechnol. 2025. PMID: 40072822 Free PMC article.
-
Evaluation of Phage Display Biopanning Strategies for the Selection of Anti-Cell Surface Receptor Antibodies.Int J Mol Sci. 2022 Jul 30;23(15):8470. doi: 10.3390/ijms23158470. Int J Mol Sci. 2022. PMID: 35955604 Free PMC article.
-
Membrane Protein Stabilization Strategies for Structural and Functional Studies.Membranes (Basel). 2021 Feb 22;11(2):155. doi: 10.3390/membranes11020155. Membranes (Basel). 2021. PMID: 33671740 Free PMC article. Review.
References
-
- Bartoccioni P., Del Rio C., Ratera M., Kowalczyk L., Baldwin J.M., Zorzano A., Quick M., Baldwin S.A., Vázquez-Ibar J.L., and Palacín M.. 2010. Role of transmembrane domain 8 in substrate selectivity and translocation of SteT, a member of the L-amino acid transporter (LAT) family. J. Biol. Chem. 285:28764–28776. 10.1074/jbc.M110.116632 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

