Functionally Similar WRKY Proteins Regulate Vacuolar Acidification in Petunia and Hair Development in Arabidopsis
- PMID: 26977085
- PMCID: PMC4826004
- DOI: 10.1105/tpc.15.00608
Functionally Similar WRKY Proteins Regulate Vacuolar Acidification in Petunia and Hair Development in Arabidopsis
Abstract
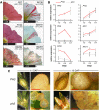
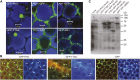
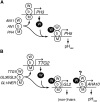
The WD40 proteins ANTHOCYANIN11 (AN11) from petunia (Petunia hybrida) and TRANSPARENT TESTA GLABRA1 (TTG1) from Arabidopsis thaliana and associated basic helix-loop-helix (bHLH) and MYB transcription factors activate a variety of differentiation processes. In petunia petals, AN11 and the bHLH protein AN1 activate, together with the MYB protein AN2, anthocyanin biosynthesis and, together with the MYB protein PH4, distinct genes, such as PH1 and PH5, that acidify the vacuole. To understand how AN1 and AN11 activate anthocyanin biosynthetic and PH genes independently, we isolated PH3. We found that PH3 is a target gene of the AN11-AN1-PH4 complex and encodes a WRKY protein that can bind to AN11 and is required, in a feed-forward loop, together with AN11-AN1-PH4 for transcription of PH5. PH3 is highly similar to TTG2, which regulates hair development, tannin accumulation, and mucilage production in Arabidopsis. Like PH3, TTG2 can bind to petunia AN11 and the Arabidopsis homolog TTG1, complement ph3 in petunia, and reactivate the PH3 target gene PH5. Our findings show that the specificity of WD40-bHLH-MYB complexes is in part determined by interacting proteins, such as PH3 and TTG2, and reveal an unanticipated similarity in the regulatory circuitry that controls petunia vacuolar acidification and Arabidopsis hair development.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures






References
-
- Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. (2011). Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65: 771–784. - PubMed
-
- Appelhagen I., Nordholt N., Seidel T., Spelt K., Koes R., Quattrochio F., Sagasser M., Weisshaar B. (2015). TRANSPARENT TESTA 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82: 840–849. - PubMed
-
- Assaad F.F., Qiu J.L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129. - PMC - PubMed
-
- Balkunde R., Bouyer D., Hülskamp M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048. - PubMed
-
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

