PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion
- PMID: 26977884
- PMCID: PMC5300038
- DOI: 10.1016/j.ccell.2016.02.013
PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion
Abstract
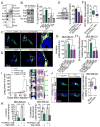
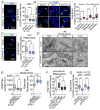
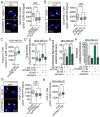
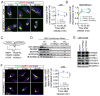
Enhanced secretion of tumorigenic effector proteins is a feature of malignant cells. The molecular mechanisms underlying this feature are poorly defined. We identify PITPNC1 as a gene amplified in a large fraction of human breast cancer and overexpressed in metastatic breast, melanoma, and colon cancers. Biochemical, molecular, and cell-biological studies reveal that PITPNC1 promotes malignant secretion by binding Golgi-resident PI4P and localizing RAB1B to the Golgi. RAB1B localization to the Golgi allows for the recruitment of GOLPH3, which facilitates Golgi extension and enhanced vesicular release. PITPNC1-mediated vesicular release drives metastasis by increasing the secretion of pro-invasive and pro-angiogenic mediators HTRA1, MMP1, FAM3C, PDGFA, and ADAM10. We establish PITPNC1 as a PI4P-binding protein that enhances vesicular secretion capacity in malignancy.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Anan K, Morisaki T, Katano M, Ikubo A, Kitsuki H, Uchiyama A, Kuroki S, Tanaka M, Torisu M. Vascular endothelial growth factor and platelet-derived growth factor are potential angiogenic and metastatic factors in human breast cancer. Surgery. 1996;119:333–339. - PubMed
-
- Bonazzi M, Spano S, Turacchio G, Cericola C, Valente C, Colanzi A, Kweon HS, Hsu VW, Polishchuck EV, Polishchuck RS, et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005;7:570–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

