Plant Tandem CCCH Zinc Finger Proteins Interact with ABA, Drought, and Stress Response Regulators in Processing-Bodies and Stress Granules
- PMID: 26978070
- PMCID: PMC4792416
- DOI: 10.1371/journal.pone.0151574
Plant Tandem CCCH Zinc Finger Proteins Interact with ABA, Drought, and Stress Response Regulators in Processing-Bodies and Stress Granules
Abstract
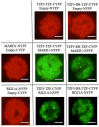
Although multiple lines of evidence have indicated that Arabidopsis thaliana Tandem CCCH Zinc Finger proteins, AtTZF4, 5 and 6 are involved in ABA, GA and phytochrome mediated seed germination responses, the interacting proteins involved in these processes are unknown. Using yeast two-hybrid screens, we have identified 35 putative AtTZF5 interacting protein partners. Among them, Mediator of ABA-Regulated Dormancy 1 (MARD1) is highly expressed in seeds and involved in ABA signal transduction, while Responsive to Dehydration 21A (RD21A) is a well-documented stress responsive protein. Co-immunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays were used to confirm that AtTZF5 can interact with MARD1 and RD21A in plant cells, and the interaction is mediated through TZF motif. In addition, AtTZF4 and 6 could also interact with MARD1 and RD21A in Y-2-H and BiFC assay, respectively. The protein-protein interactions apparently take place in processing bodies (PBs) and stress granules (SGs), because AtTZF5, MARD1 and RD21A could interact and co-localize with each other and they all can co-localize with the same PB and SG markers in plant cells.
Conflict of interest statement
Figures






References
-
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, et al. Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant. 2013;6(1752–9867 (Electronic)):287–300. - PubMed
-
- Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14(0890–9369 (Print)):1027–47. - PubMed
-
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Bichimica et Biophysica Acta. 2011;1808:912–24. - PubMed
-
- Jackman MR, Pines JN. Cyclins and the G2/M transition. Cancer Surv. 1997;29(0261–2429 (Print)):47–73. - PubMed
-
- Jeffrey PD, Russo AA, Polyak K, Gibbs H, Hurwitz J, Massague J, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376(0028–0836 (Print)):313–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

