Dynamic interplay between locus-specific DNA methylation and hydroxymethylation regulates distinct biological pathways in prostate carcinogenesis
- PMID: 26981160
- PMCID: PMC4791926
- DOI: 10.1186/s13148-016-0195-4
Dynamic interplay between locus-specific DNA methylation and hydroxymethylation regulates distinct biological pathways in prostate carcinogenesis
Abstract
Background: Despite the significant global loss of DNA hydroxymethylation marks in prostate cancer tissues, the locus-specific role of hydroxymethylation in prostate tumorigenesis is unknown. We characterized hydroxymethylation and methylation marks by performing whole-genome next-generation sequencing in representative normal and prostate cancer-derived cell lines in order to determine functional pathways and key genes regulated by these epigenomic modifications in cancer.
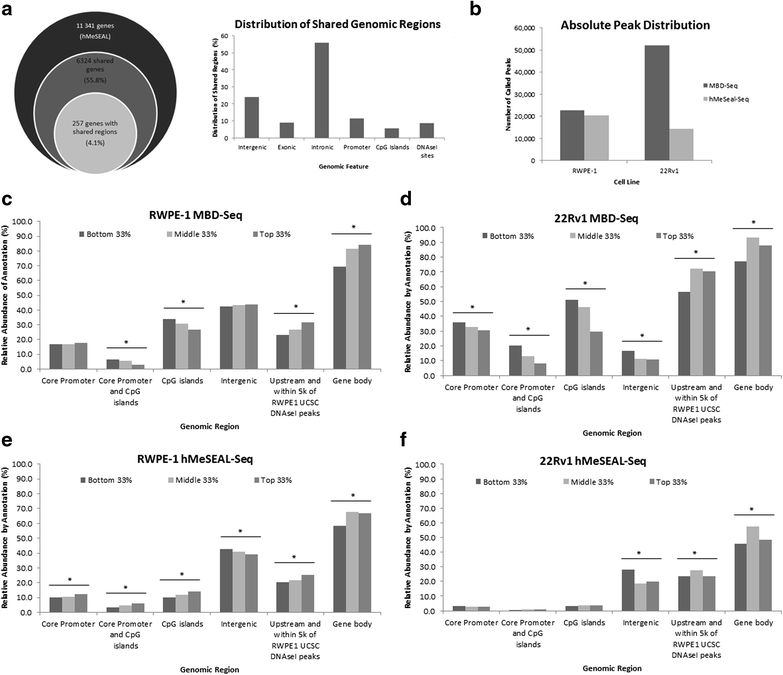
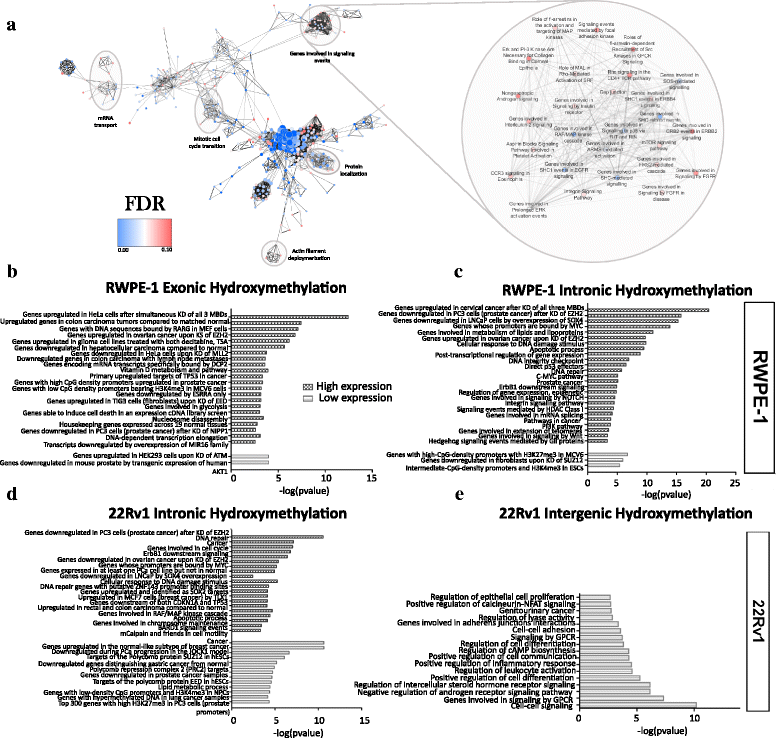
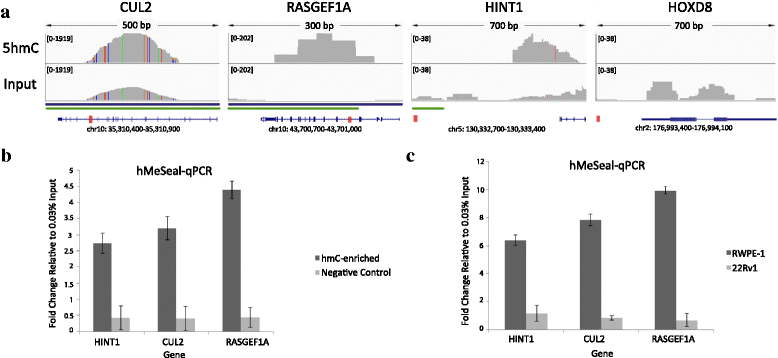
Results: Our cell line model shows disruption of hydroxymethylation distribution in cancer, with global loss and highly specific gain in promoter and CpG island regions. Significantly, we observed locus-specific retention of hydroxymethylation marks in specific intronic and intergenic regions which may play a novel role in the regulation of gene expression in critical functional pathways, such as BARD1 signaling and steroid hormone receptor signaling in cancer. We confirm a modest correlation of hydroxymethylation with expression in intragenic regions in prostate cancer, while identifying an original role for intergenic hydroxymethylation in differentially expressed regulatory pathways in cancer. We also demonstrate a successful strategy for the identification and validation of key candidate genes from differentially regulated biological pathways in prostate cancer.
Conclusions: Our results indicate a distinct function for aberrant hydroxymethylation within each genomic feature in cancer, suggesting a specific and complex role for the deregulation of hydroxymethylation in tumorigenesis, similar to methylation. Subsequently, our characterization of key cellular pathways exhibiting dynamic enrichment patterns for methylation and hydroxymethylation marks may allow us to identify differentially epigenetically modified target genes implicated in prostate cancer tumorigenesis.
Keywords: 5-Hydroxymethylcytosine; 5-Methylcytosine; Cell lines; Integrative analysis; Prostate cancer; Whole-genome.
Figures


References
-
- Van Vlodrop IJ, Niessen HE, Derks S, Baldewijns MM, van Criekinge W, Herman JG, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clin Cancer Res. 2011 - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

