Prevalence of the C-terminal truncations of NS1 in avian influenza A viruses and effect on virulence and replication of a highly pathogenic H7N1 virus in chickens
- PMID: 26981790
- PMCID: PMC5026787
- DOI: 10.1080/21505594.2016.1159367
Prevalence of the C-terminal truncations of NS1 in avian influenza A viruses and effect on virulence and replication of a highly pathogenic H7N1 virus in chickens
Abstract
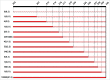
Highly pathogenic (HP) avian influenza viruses (AIV) evolve from low pathogenic (LP) precursors after circulation in poultry by reassortment and/or single mutations in different gene segments including that encoding NS1. The carboxyl terminal end (CTE) of NS1 exhibits deletions between amino acid 202 and 230 with still unknown impact on virulence of AIV in chickens. In this study, NS1 protein sequences of all AIV subtypes in birds from 1902 to 2015 were analyzed to study the prevalence and distribution of CTE truncation (ΔCTE). Thirteen different ΔCTE forms were observed in NS1 proteins from 11 HA and 8 NA subtypes with high prevalences in H9, H7, H6 and H10 and N9, N2, N6 and N1 subtypes particularly in chickens and minor poultry species. With 88% NS217 lacking amino acids 218-230 was the most common ΔCTE form followed by NS224 (3.6%). NS217 was found in 10 and 8 different HA and NA subtypes, respectively, whereas NS224 was detected exclusively in the Italian HPAIV H7N1 suggesting relevance for virulence. To test this assumption, 3 recombinant HPAIV H7N1 were constructed carrying wild-type HP NS1 (Hp-NS224), NS1 with extended CTE (Hp-NS230) or NS1 from LPAIV H7N1 (Hp-NSLp), and tested in-vitro and in-vivo. Extension of CTE in Hp NS1 significantly decreased virus replication in chicken embryo kidney cells. Truncation in the NS1 decreased the tropism of Hp-NS224 to the endothelium, central nervous system and respiratory tract epithelium without significant difference in virulence in chickens. This study described the variable forms of ΔCTE in NS1 and indicated that CTE is not an essential virulence determinant particularly for the Italian HPAIV H7N1 but may be a host-adaptation marker required for efficient virus replication.
Keywords: NS1; adaptation; avian influenza virus; chickens; deletion; high pathogenicity; virulence.
Figures


Comment in
-
Stop-codon variations in non-structural protein NS1 of avian influenza viruses.Virulence. 2016 Jul 3;7(5):498-501. doi: 10.1080/21505594.2016.1175802. Epub 2016 Apr 8. Virulence. 2016. PMID: 27058119 Free PMC article. No abstract available.
References
-
- Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol 2000; 74:3-13; PMID:10799774; http://dx.doi.org/ 10.1016/S0378-1135(00)00160-7 - DOI - PubMed
-
- Khaliq Z, Leijon M, Belak S, Komorowski J. A complete map of potential pathogenicity markers of avian influenza virus subtype H5 predicted from 11 expressed proteins. BMC Microbiol 2015; 15:128; PMID:26112351; http://dx.doi.org/ 10.1186/s12866-015-0465-x - DOI - PMC - PubMed
-
- Imai H, Shinya K, Takano R, Kiso M, Muramoto Y, Sakabe S, Murakami S, Ito M, Yamada S, Le MT, et al.. The HA and NS genes of human H5N1 influenza A virus contribute to high virulence in ferrets. PLoS Pathogens 2010; 6:e1001106; PMID:20862325; http://dx.doi.org/ 10.1371/journal.ppat.1001106 - DOI - PMC - PubMed
-
- Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, Chen H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol 2006; 80:11115-23; PMID:16971424; PMID:16971424; http://dx.doi.org/ 10.1128/JVI.00993-06 - DOI - PMC - PubMed
-
- Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 2014; 88:12146-51; PMID:25078692; http://dx.doi.org/ 10.1128/JVI.01567-14 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
