Evaluation of Viremia Frequencies of a Novel Human Pegivirus by Using Bioinformatic Screening and PCR
- PMID: 26982117
- PMCID: PMC4806942
- DOI: 10.3201/eid2204.151812
Evaluation of Viremia Frequencies of a Novel Human Pegivirus by Using Bioinformatic Screening and PCR
Abstract
Next-generation sequencing has critical applications in virus discovery, diagnostics, and environmental surveillance. We used metagenomic sequence libraries for retrospective screening of plasma samples for the recently discovered human hepegivirus 1 (HHpgV-1). From a cohort of 150 hepatitis C virus (HCV)-positive case-patients, we identified 2 persons with HHpgV-1 viremia and a high frequency of human pegivirus (HPgV) viremia (14%). Detection of HHpgV-1 and HPgV was concordant with parallel PCR-based screening using conserved primers matching groups 1 (HPgV) and 2 (HHPgV-1) nonstructural 3 region sequences. PCR identified 1 HHPgV-1-positive person with viremia from a group of 195 persons with hemophilia who had been exposed to nonvirally inactivated factor VII/IX; 18 (9%) were HPgV-positive. Relative to HCV and HPgV, active infections with HHpgV-1 were infrequently detected in blood, even in groups that had substantial parenteral exposure. Our findings are consistent with lower transmissibility or higher rates of virus clearance for HHpgV-1 than for other bloodborne human flaviviruses.
Keywords: Flaviviridae; HCV; HHpgV-1; Hepacivirus; Hepegivirus; Pegivirus; bloodborne pathogens; hemophilia; parenteral; persons who inject drugs; sexually transmitted disease; transfusion; virus persistence; viruses.
Figures
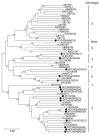
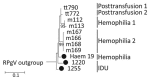
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

