Evolution of KaiC-Dependent Timekeepers: A Proto-circadian Timing Mechanism Confers Adaptive Fitness in the Purple Bacterium Rhodopseudomonas palustris
- PMID: 26982486
- PMCID: PMC4794148
- DOI: 10.1371/journal.pgen.1005922
Evolution of KaiC-Dependent Timekeepers: A Proto-circadian Timing Mechanism Confers Adaptive Fitness in the Purple Bacterium Rhodopseudomonas palustris
Abstract
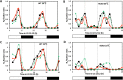
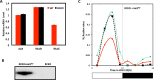
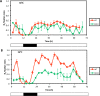
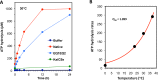
Circadian (daily) rhythms are a fundamental and ubiquitous property of eukaryotic organisms. However, cyanobacteria are the only prokaryotic group for which bona fide circadian properties have been persuasively documented, even though homologs of the cyanobacterial kaiABC central clock genes are distributed widely among Eubacteria and Archaea. We report the purple non-sulfur bacterium Rhodopseudomonas palustris (that harbors homologs of kaiB and kaiC) only poorly sustains rhythmicity in constant conditions-a defining characteristic of circadian rhythms. Moreover, the biochemical characteristics of the Rhodopseudomonas homolog of the KaiC protein in vivo and in vitro are different from those of cyanobacterial KaiC. Nevertheless, R. palustris cells exhibit adaptive kaiC-dependent growth enhancement in 24-h cyclic environments, but not under non-natural constant conditions. Therefore, our data indicate that Rhodopseudomonas does not have a classical circadian rhythm, but a novel timekeeping mechanism that does not sustain itself in constant conditions. These results question the adaptive value of self-sustained oscillatory capability for daily timekeepers and establish new criteria for circadian-like systems that are based on adaptive properties (i.e., fitness enhancement in rhythmic environments), rather than upon observations of persisting rhythms in constant conditions. We propose that the Rhodopseudomonas system is a "proto" circadian timekeeper, as in an ancestral system that is based on KaiC and KaiB proteins and includes some, but not necessarily all, of the canonical properties of circadian clocks. These data indicate reasonable intermediate steps by which bona fide circadian systems evolved in simple organisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Dunlap JC Chronobiology: Biological timekeeping Sunderland, Mass.: Sinauer; 2004.
-
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14: 1481–1486. - PubMed
-
- Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996; 21: 5–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

