Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod
- PMID: 26983361
- PMCID: PMC4794648
- DOI: 10.1038/srep23246
Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod
Abstract
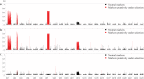
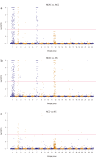
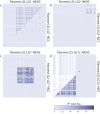
Identification of genome-wide patterns of divergence provides insight on how genomes are influenced by selection and can reveal the potential for local adaptation in spatially structured populations. In Atlantic cod - historically a major marine resource - Northeast-Arctic- and Norwegian coastal cod are recognized by fundamental differences in migratory and non-migratory behavior, respectively. However, the genomic architecture underlying such behavioral ecotypes is unclear. Here, we have analyzed more than 8.000 polymorphic SNPs distributed throughout all 23 linkage groups and show that loci putatively under selection are localized within three distinct genomic regions, each of several megabases long, covering approximately 4% of the Atlantic cod genome. These regions likely represent genomic inversions. The frequency of these distinct regions differ markedly between the ecotypes, spawning in the vicinity of each other, which contrasts with the low level of divergence in the rest of the genome. The observed patterns strongly suggest that these chromosomal rearrangements are instrumental in local adaptation and separation of Atlantic cod populations, leaving footprints of large genomic regions under selection. Our findings demonstrate the power of using genomic information in further understanding the population dynamics and defining management units in one of the world's most economically important marine resources.
Figures



References
-
- Wu C.-I. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865 (2001).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

